Widespread presence of human BOULE homologs among animals and conservation of their ancient reproductive function
- PMID: 20657660
- PMCID: PMC2904765
- DOI: 10.1371/journal.pgen.1001022
Widespread presence of human BOULE homologs among animals and conservation of their ancient reproductive function
Abstract
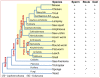
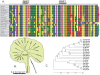
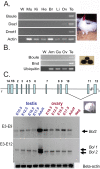
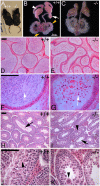
Sex-specific traits that lead to the production of dimorphic gametes, sperm in males and eggs in females, are fundamental for sexual reproduction and accordingly widespread among animals. Yet the sex-biased genes that underlie these sex-specific traits are under strong selective pressure, and as a result of adaptive evolution they often become divergent. Indeed out of hundreds of male or female fertility genes identified in diverse organisms, only a very small number of them are implicated specifically in reproduction in more than one lineage. Few genes have exhibited a sex-biased, reproductive-specific requirement beyond a given phylum, raising the question of whether any sex-specific gametogenesis factors could be conserved and whether gametogenesis might have evolved multiple times. Here we describe a metazoan origin of a conserved human reproductive protein, BOULE, and its prevalence from primitive basal metazoans to chordates. We found that BOULE homologs are present in the genomes of representative species of each of the major lineages of metazoans and exhibit reproductive-specific expression in all species examined, with a preponderance of male-biased expression. Examination of Boule evolution within insect and mammalian lineages revealed little evidence for accelerated evolution, unlike most reproductive genes. Instead, purifying selection was the major force behind Boule evolution. Furthermore, loss of function of mammalian Boule resulted in male-specific infertility and a global arrest of sperm development remarkably similar to the phenotype in an insect boule mutation. This work demonstrates the conservation of a reproductive protein throughout eumetazoa, its predominant testis-biased expression in diverse bilaterian species, and conservation of a male gametogenic requirement in mice. This shows an ancient gametogenesis requirement for Boule among Bilateria and supports a model of a common origin of spermatogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule.Hum Mol Genet. 2010 Jun 15;19(12):2360-9. doi: 10.1093/hmg/ddq109. Epub 2010 Mar 24. Hum Mol Genet. 2010. PMID: 20335278 Free PMC article.
-
Human BOULE gene rescues meiotic defects in infertile flies.Hum Mol Genet. 2003 Jan 15;12(2):169-75. doi: 10.1093/hmg/ddg017. Hum Mol Genet. 2003. PMID: 12499397
-
Differential conservation and divergence of fertility genes boule and dazl in the rainbow trout.PLoS One. 2011 Jan 6;6(1):e15910. doi: 10.1371/journal.pone.0015910. PLoS One. 2011. PMID: 21253610 Free PMC article.
-
LncRNA, a new component of expanding RNA-protein regulatory network important for animal sperm development.Semin Cell Dev Biol. 2016 Nov;59:110-117. doi: 10.1016/j.semcdb.2016.06.013. Epub 2016 Jun 21. Semin Cell Dev Biol. 2016. PMID: 27345292 Review.
-
Role of the DAZ genes in male fertility.Reprod Biomed Online. 2005 Jan;10(1):72-80. doi: 10.1016/s1472-6483(10)60806-1. Reprod Biomed Online. 2005. PMID: 15705297 Review.
Cited by
-
Genome-wide transcriptome profiling and spatial expression analyses identify signals and switches of development in tapeworms.Evodevo. 2018 Nov 9;9:21. doi: 10.1186/s13227-018-0110-5. eCollection 2018. Evodevo. 2018. PMID: 30455861 Free PMC article.
-
Epigenetic regulation of bovine spermatogenic cell-specific gene boule.PLoS One. 2015 Jun 1;10(6):e0128250. doi: 10.1371/journal.pone.0128250. eCollection 2015. PLoS One. 2015. PMID: 26030766 Free PMC article.
-
The conserved genetic program of male germ cells uncovers ancient regulators of human spermatogenesis.Elife. 2024 Oct 10;13:RP95774. doi: 10.7554/eLife.95774. Elife. 2024. PMID: 39388236 Free PMC article.
-
The roles of the DAZ family in spermatogenesis: More than just translation?Spermatogenesis. 2011 Jan 1;1(1):36-46. doi: 10.4161/spmg.1.1.14659. Spermatogenesis. 2011. PMID: 22523742 Free PMC article.
-
DAZL is a master translational regulator of murine spermatogenesis.Natl Sci Rev. 2019 May;6(3):455-468. doi: 10.1093/nsr/nwy163. Epub 2018 Dec 28. Natl Sci Rev. 2019. PMID: 31355046 Free PMC article.
References
-
- Ramesh MA, Malik SB, Logsdon JM., Jr A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol. 2005;15:185–191. - PubMed
-
- Villeneuve AM, Hillers KJ. Whence meiosis? Cell. 2001;106:647–650. - PubMed
-
- Charlesworth B. The evolution of sex chromosome. Science. 1991;251:1030–1033. - PubMed
-
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. - PubMed
-
- Wyckoff G, Wang W, Wu C-I. Rapid evolution of reproductive genes in the descent of man. Nature. 2000;403:304–309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

