Organization of cellular receptors into a nanoscale junction during HIV-1 adhesion
- PMID: 20657663
- PMCID: PMC2904768
- DOI: 10.1371/journal.pcbi.1000855
Organization of cellular receptors into a nanoscale junction during HIV-1 adhesion
Abstract
The fusion of the human immunodeficiency virus type 1 (HIV-1) with its host cell is the target for new antiretroviral therapies. Viral particles interact with the flexible plasma membrane via viral surface protein gp120 which binds its primary cellular receptor CD4 and subsequently the coreceptor CCR5. However, whether and how these receptors become organized at the adhesive junction between cell and virion are unknown. Here, stochastic modeling predicts that, regarding binding to gp120, cellular receptors CD4 and CCR5 form an organized, ring-like, nanoscale structure beneath the virion, which locally deforms the plasma membrane. This organized adhesive junction between cell and virion, which we name the viral junction, is reminiscent of the well-characterized immunological synapse, albeit at much smaller length scales. The formation of an organized viral junction under multiple physiopathologically relevant conditions may represent a novel intermediate step in productive infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
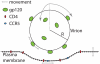
 = 50nm, and was populated by gp120 trimers (green) capable of binding up to 3 CD4 and 3 CCR5 molecules. All entities were prohibited from diffusing through one another. The viral particle was allowed to move in 3 dimensions as well as rotate about its center, proteins were allowed to move in 2 dimensions along their respective surfaces and discrete plasma membrane points were allowed to move in 1 dimension either up or down. Movement directions within the plane here are shown with grey arrows.
= 50nm, and was populated by gp120 trimers (green) capable of binding up to 3 CD4 and 3 CCR5 molecules. All entities were prohibited from diffusing through one another. The viral particle was allowed to move in 3 dimensions as well as rotate about its center, proteins were allowed to move in 2 dimensions along their respective surfaces and discrete plasma membrane points were allowed to move in 1 dimension either up or down. Movement directions within the plane here are shown with grey arrows.
 , the change in position of viral center from initial cell contact) decreases, approaching a steady state. (G) A typical realization of the viral junction including a rigid sphere above a flexible plasma membrane with CD4/CCR5 bond locations projected beneath. (H) The plasma membrane of the system deforms and approaches the curvature of the partially engulfed virion at steady state. Shown here is a radial profile of the plasma membrane i.e., its vertical displacement as a function of the radial distance from the viral center. Simulations were performed with 15 gp120 trimers per virion and eight such simulations were averaged to produce the results illustrated here.
, the change in position of viral center from initial cell contact) decreases, approaching a steady state. (G) A typical realization of the viral junction including a rigid sphere above a flexible plasma membrane with CD4/CCR5 bond locations projected beneath. (H) The plasma membrane of the system deforms and approaches the curvature of the partially engulfed virion at steady state. Shown here is a radial profile of the plasma membrane i.e., its vertical displacement as a function of the radial distance from the viral center. Simulations were performed with 15 gp120 trimers per virion and eight such simulations were averaged to produce the results illustrated here.


 is the change in position of viral center from initial cell contact. Conditions correspond to panels A–D, as indicated. (H) The steady state profile of the plasma membrane (the x-axis is measured in nm from the viral center, while the y-axis is the average variation of the membrane height from initialization of the system). Conditions correspond to panels A–D, as indicated. Simulations were performed with 15 gp120 trimers per virion and eight such simulations were averaged to produce the results displayed here.
is the change in position of viral center from initial cell contact. Conditions correspond to panels A–D, as indicated. (H) The steady state profile of the plasma membrane (the x-axis is measured in nm from the viral center, while the y-axis is the average variation of the membrane height from initialization of the system). Conditions correspond to panels A–D, as indicated. Simulations were performed with 15 gp120 trimers per virion and eight such simulations were averaged to produce the results displayed here.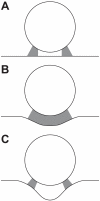

 is the change in position of viral center from initial cell contact. Conditions correspond to panels A–D, as indicated. (H) The steady state profile of the plasma membrane (the x-axis is measured in nm from the viral center, while the y-axis is the average variation of the membrane height from initialization of the system). Conditions correspond to panels A–D, as indicated. Simulations were performed with 15 gp120 trimers per virion and eight such simulations were averaged to produce the results displayed here.
is the change in position of viral center from initial cell contact. Conditions correspond to panels A–D, as indicated. (H) The steady state profile of the plasma membrane (the x-axis is measured in nm from the viral center, while the y-axis is the average variation of the membrane height from initialization of the system). Conditions correspond to panels A–D, as indicated. Simulations were performed with 15 gp120 trimers per virion and eight such simulations were averaged to produce the results displayed here.Similar articles
-
Binding thermodynamics of the N-terminal peptide of the CCR5 coreceptor to HIV-1 envelope glycoprotein gp120.Biochemistry. 2009 Feb 3;48(4):779-85. doi: 10.1021/bi8021476. Biochemistry. 2009. PMID: 19170639 Free PMC article.
-
Activation and Inactivation of Primary Human Immunodeficiency Virus Envelope Glycoprotein Trimers by CD4-Mimetic Compounds.J Virol. 2017 Jan 18;91(3):e01880-16. doi: 10.1128/JVI.01880-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881646 Free PMC article.
-
Single-Molecule Super-Resolution Imaging of T-Cell Plasma Membrane CD4 Redistribution upon HIV-1 Binding.Viruses. 2021 Jan 19;13(1):142. doi: 10.3390/v13010142. Viruses. 2021. PMID: 33478139 Free PMC article.
-
Membrane organization of virus and target cell plays a role in HIV entry.Biochimie. 2014 Dec;107 Pt A:22-7. doi: 10.1016/j.biochi.2014.08.015. Epub 2014 Sep 1. Biochimie. 2014. PMID: 25193376 Free PMC article. Review.
-
HIV-1 attachment: another look.Trends Microbiol. 1999 Apr;7(4):144-9. doi: 10.1016/s0966-842x(99)01474-2. Trends Microbiol. 1999. PMID: 10217828 Review.
Cited by
-
Multiscale perspectives of virus entry via endocytosis.Virol J. 2013 Jun 5;10:177. doi: 10.1186/1743-422X-10-177. Virol J. 2013. PMID: 23734580 Free PMC article. Review.
-
Catching HIV 'in the act' with 3D electron microscopy.Trends Microbiol. 2013 Aug;21(8):397-404. doi: 10.1016/j.tim.2013.06.004. Epub 2013 Jul 11. Trends Microbiol. 2013. PMID: 23850373 Free PMC article. Review.
-
Cell Surface Mechanochemistry and the Determinants of Bleb Formation, Healing, and Travel Velocity.Biophys J. 2016 Apr 12;110(7):1636-1647. doi: 10.1016/j.bpj.2016.03.008. Biophys J. 2016. PMID: 27074688 Free PMC article.
-
Catch Bonds at T Cell Interfaces: Impact of Surface Reorganization and Membrane Fluctuations.Biophys J. 2017 Jul 11;113(1):120-131. doi: 10.1016/j.bpj.2017.05.023. Biophys J. 2017. PMID: 28700910 Free PMC article.
-
HIV entry and envelope glycoprotein-mediated fusion.J Biol Chem. 2012 Nov 30;287(49):40841-9. doi: 10.1074/jbc.R112.406272. Epub 2012 Oct 5. J Biol Chem. 2012. PMID: 23043104 Free PMC article. Review.
References
-
- Clapham PR, McKnight A. Cell surface receptors, virus entry and tropism of primate lentiviruses. J Gen Virol. 2002;83:1809–1829. - PubMed
-
- Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. - PubMed
-
- Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. - PubMed
-
- Dustin ML. T-cell activation through immunological synapses and kinapses. Immunol Rev. 2008;221:77–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

