Mouse cytoplasmic dynein intermediate chains: identification of new isoforms, alternative splicing and tissue distribution of transcripts
- PMID: 20657784
- PMCID: PMC2908135
- DOI: 10.1371/journal.pone.0011682
Mouse cytoplasmic dynein intermediate chains: identification of new isoforms, alternative splicing and tissue distribution of transcripts
Erratum in
- PLoS One. 2010;5(10). doi: 10.1371/annotation/59badad8-6e55-46f8-8bf1-7a8a957bc68e doi: 10.1371/annotation/59badad8-6e55-46f8-8bf1-7a8a957bc68e
Abstract
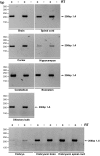
Background: Intracellular transport of cargoes including organelles, vesicles, signalling molecules, protein complexes, and RNAs, is essential for normal function of eukaryotic cells. The cytoplasmic dynein complex is an important motor that moves cargos along microtubule tracks within the cell. In mammals this multiprotein complex includes dynein intermediate chains 1 and 2 which are encoded by two genes, Dync1i1 and Dync1i2. These proteins are involved in dynein cargo binding and dynein complexes with different intermediate chains bind to specific cargoes, although the mechanisms to achieve this are not known. The DYNC1I1 and DYNC1I2 proteins are translated from different splice isoforms, and specific forms of each protein are essential for the function of different dynein complexes in neurons.
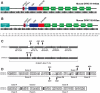
Methodology/principal findings: Here we have undertaken a systematic survey of the dynein intermediate chain splice isoforms in mouse, basing our study on mRNA expression patterns in a range of tissues, and on bioinformatics analysis of mouse, rat and human genomic and cDNA sequences. We found a complex pattern of alternative splicing of both dynein intermediate chain genes, with maximum complexity in the embryonic and adult nervous system. We have found novel transcripts, including some with orthologues in human and rat, and a new promoter and alternative non-coding exon 1 for Dync1i2.
Conclusions/significance: These data, including the cloned isoforms will be essential for understanding the role of intermediate chains in the cytoplasmic dynein complex, particularly their role in cargo binding within individual tissues including different brain regions.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

