Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation
- PMID: 20657825
- PMCID: PMC2904796
- DOI: 10.1371/journal.pgen.1001024
Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation
Abstract
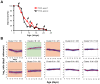
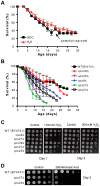
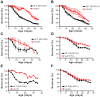
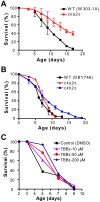
The study of the chronological life span of Saccharomyces cerevisiae, which measures the survival of populations of non-dividing yeast, has resulted in the identification of homologous genes and pathways that promote aging in organisms ranging from yeast to mammals. Using a competitive genome-wide approach, we performed a screen of a complete set of approximately 4,800 viable deletion mutants to identify genes that either increase or decrease chronological life span. Half of the putative short-/long-lived mutants retested from the primary screen were confirmed, demonstrating the utility of our approach. Deletion of genes involved in vacuolar protein sorting, autophagy, and mitochondrial function shortened life span, confirming that respiration and degradation processes are essential for long-term survival. Among the genes whose deletion significantly extended life span are ACB1, CKA2, and TRM9, implicated in fatty acid transport and biosynthesis, cell signaling, and tRNA methylation, respectively. Deletion of these genes conferred heat-shock resistance, supporting the link between life span extension and cellular protection observed in several model organisms. The high degree of conservation of these novel yeast longevity determinants in other species raises the possibility that their role in senescence might be conserved.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Endosomal protein sorting and autophagy genes contribute to the regulation of yeast life span.Autophagy. 2010 Nov;6(8):1227-8. doi: 10.4161/auto.6.8.13850. Epub 2010 Nov 16. Autophagy. 2010. PMID: 20953148
-
A genomic analysis of chronological longevity factors in budding yeast.Cell Cycle. 2011 May 1;10(9):1385-96. doi: 10.4161/cc.10.9.15464. Epub 2011 May 1. Cell Cycle. 2011. PMID: 21447998 Free PMC article.
-
Gene-nutrient interaction markedly influences yeast chronological lifespan.Exp Gerontol. 2016 Dec 15;86:113-123. doi: 10.1016/j.exger.2016.04.012. Epub 2016 Apr 25. Exp Gerontol. 2016. PMID: 27125759 Free PMC article.
-
Genome-wide analysis of yeast aging.Subcell Biochem. 2012;57:251-89. doi: 10.1007/978-94-007-2561-4_12. Subcell Biochem. 2012. PMID: 22094426 Review.
-
Chronological aging-induced apoptosis in yeast.Biochim Biophys Acta. 2008 Jul;1783(7):1280-5. doi: 10.1016/j.bbamcr.2008.03.017. Epub 2008 Apr 10. Biochim Biophys Acta. 2008. PMID: 18445486 Free PMC article. Review.
Cited by
-
Cryptococcus neoformans constitutes an ideal model organism to unravel the contribution of cellular aging to the virulence of chronic infections.Curr Opin Microbiol. 2013 Aug;16(4):391-7. doi: 10.1016/j.mib.2013.03.011. Epub 2013 Apr 27. Curr Opin Microbiol. 2013. PMID: 23631868 Free PMC article. Review.
-
Genome-wide screens in yeast models towards understanding chronological lifespan regulation.Brief Funct Genomics. 2022 Jan 25;21(1):4-12. doi: 10.1093/bfgp/elab011. Brief Funct Genomics. 2022. PMID: 33728458 Free PMC article.
-
Yeast colonies: a model for studies of aging, environmental adaptation, and longevity.Oxid Med Cell Longev. 2012;2012:601836. doi: 10.1155/2012/601836. Epub 2012 Aug 13. Oxid Med Cell Longev. 2012. PMID: 22928081 Free PMC article. Review.
-
Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species.Aging Cell. 2019 Dec;18(6):e13034. doi: 10.1111/acel.13034. Epub 2019 Aug 28. Aging Cell. 2019. PMID: 31460700 Free PMC article. Review.
-
Aging and cell death in the other yeasts, Schizosaccharomyces pombe and Candida albicans.FEMS Yeast Res. 2014 Feb;14(1):119-35. doi: 10.1111/1567-1364.12113. Epub 2013 Nov 8. FEMS Yeast Res. 2014. PMID: 24205865 Free PMC article. Review.
References
-
- Longo VD, Finch CE. Evolutionary medicine: from dwarf model systems to healthy centenarians. Science. 2003;299:1342–1346. - PubMed
-
- Kenyon C. A conserved regulatory system for aging. Cell. 2001;105:165–168. - PubMed
-
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Methods Mol Biol. 2007;371:89–95. - PubMed
-
- Mortimer RK. Life span of individual yeast cells. Nature. 1959;183:1751–1752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

