C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues
- PMID: 20659444
- PMCID: PMC2955628
- DOI: 10.1016/j.ydbio.2010.07.020
C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues
Abstract
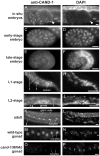
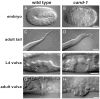
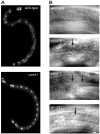
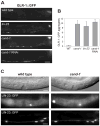
Cullin-RING ubiquitin ligases (CRLs) are critical regulators of multiple developmental and cellular processes in eukaryotes. CAND1 is a biochemical inhibitor of CRLs, yet has been shown to promote CRL activity in plant and mammalian cells. Here we analyze CAND1 function in the context of a developing metazoan organism. Caenorhabditis elegans CAND-1 is capable of binding to all of the cullins, and we show that it physically interacts with CUL-2 and CUL-4 in vivo. The covalent attachment of the ubiquitin-like protein Nedd8 is required for cullin activity in animals and plants. In cand-1 mutants, the levels of the neddylated isoforms of CUL-2 and CUL-4 are increased, indicating that CAND-1 is a negative regulator of cullin neddylation. cand-1 mutants are hypersensitive to the partial loss of cullin activity, suggesting that CAND-1 facilitates CRL functions. cand-1 mutants exhibit impenetrant phenotypes, including developmental arrest, morphological defects of the vulva and tail, and reduced fecundity. cand-1 mutants share with cul-1 and lin-23 mutants the phenotypes of supernumerary seam cell divisions, defective alae formation, and the accumulation of the SCF(LIN-23) target the glutamate receptor GLR-1. The observation that cand-1 mutants have phenotypes associated with the loss of the SCF(LIN-23) complex, but lack phenotypes associated with other specific CRL complexes, suggests that CAND-1 is differentially required for the activity of distinct CRL complexes.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


References
-
- Chew EH, Hagen T. Substrate-mediated regulation of cullin neddylation. J Biol Chem. 2007;282:17032–40. - PubMed
-
- Chew EH, Poobalasingam T, Hawkey CJ, Hagen T. Characterization of cullin-based E3 ubiquitin ligases in intact mammalian cells--evidence for cullin dimerization. Cell Signal. 2007;19:1071–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

