Temporal specification of blood progenitors from mouse embryonic stem cells and induced pluripotent stem cells
- PMID: 20659975
- PMCID: PMC2938916
- DOI: 10.1242/dev.042119
Temporal specification of blood progenitors from mouse embryonic stem cells and induced pluripotent stem cells
Abstract
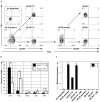
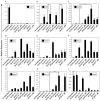
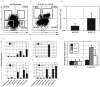
The efficient and reproducible generation of differentiated progenitors from pluripotent stem cells requires the recapitulation of appropriate developmental stages and pathways. Here, we have used the combination of activin A, BMP4 and VEGF under serum-free conditions to induce hematopoietic differentiation from both embryonic and induced pluripotent stem cells, with the aim of modeling the primary sites of embryonic hematopoiesis. We identified two distinct Flk1-positive hematopoietic populations that can be isolated based on temporal patterns of emergence. The earliest arising population displays characteristics of yolk sac hematopoiesis, whereas a late developing Flk1-positive population appears to reflect the para-aortic splanchnopleura hematopoietic program, as it has reduced primitive erythroid capacity and substantially enhanced myeloid and lymphoid potential compared with the earlier wave. These differences between the two populations are accompanied by differences in the expression of Sox17 and Hoxb4, as well as in the cell surface markers AA4.1 and CD41. Together, these findings support the interpretation that the two populations are representative of the early sites of mammalian hematopoiesis.
Figures
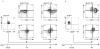


References
-
- Boisset J. C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., Robin C. (2010). In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116-120 - PubMed
-
- Cho S. K., Bourdeau A., Letarte M., Zuniga-Pflucker J. C. (2001). Expression and function of CD105 during the onset of hematopoiesis from Flk1(+) precursors. Blood 98, 3635-3642 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

