A molecular clamp ensures allosteric coordination of peptidyltransfer and ligand binding to the ribosomal A-site
- PMID: 20660012
- PMCID: PMC2995063
- DOI: 10.1093/nar/gkq641
A molecular clamp ensures allosteric coordination of peptidyltransfer and ligand binding to the ribosomal A-site
Abstract
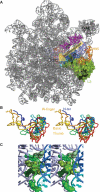
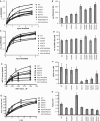
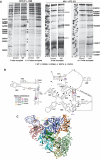
Although the ribosome is mainly comprised of rRNA and many of its critical functions occur through RNA-RNA interactions, distinct domains of ribosomal proteins also participate in switching the ribosome between different conformational/functional states. Prior studies demonstrated that two extended domains of ribosomal protein L3 form an allosteric switch between the pre- and post-translocational states. Missing was an explanation for how the movements of these domains are communicated among the ribosome's functional centers. Here, a third domain of L3 called the basic thumb, that protrudes roughly perpendicular from the W-finger and is nestled in the center of a cagelike structure formed by elements from three separate domains of the large subunit rRNA is investigated. Mutagenesis of basically charged amino acids of the basic thumb to alanines followed by detailed analyses suggests that it acts as a molecular clamp, playing a role in allosterically communicating the ribosome's tRNA occupancy status to the elongation factor binding region and the peptidyltransferase center, facilitating coordination of their functions through the elongation cycle. The observation that these mutations affected translational fidelity, virus propagation and cell growth demonstrates how small structural changes at the atomic scale can propagate outward to broadly impact the biology of cell.
Figures

Similar articles
-
Ribosomal protein L3 functions as a 'rocker switch' to aid in coordinating of large subunit-associated functions in eukaryotes and Archaea.Nucleic Acids Res. 2008 Nov;36(19):6175-86. doi: 10.1093/nar/gkn642. Epub 2008 Oct 2. Nucleic Acids Res. 2008. PMID: 18832371 Free PMC article.
-
Eukaryotic rpL10 drives ribosomal rotation.Nucleic Acids Res. 2014 Feb;42(3):2049-63. doi: 10.1093/nar/gkt1107. Epub 2013 Nov 8. Nucleic Acids Res. 2014. PMID: 24214990 Free PMC article.
-
Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis.Mol Cell Biol. 2005 Dec;25(24):10863-74. doi: 10.1128/MCB.25.24.10863-10874.2005. Mol Cell Biol. 2005. PMID: 16314511 Free PMC article.
-
Mechanism of peptide bond formation on the ribosome.Q Rev Biophys. 2006 Aug;39(3):203-25. doi: 10.1017/S003358350600429X. Epub 2006 Aug 8. Q Rev Biophys. 2006. PMID: 16893477 Review.
-
[Mechanism of peptide bond formation on the ribosome--controversions].Postepy Biochem. 2006;52(2):166-72. Postepy Biochem. 2006. PMID: 17078506 Review. Polish.
Cited by
-
Protein methylation at the surface and buried deep: thinking outside the histone box.Trends Biochem Sci. 2013 May;38(5):243-52. doi: 10.1016/j.tibs.2013.02.004. Epub 2013 Mar 13. Trends Biochem Sci. 2013. PMID: 23490039 Free PMC article. Review.
-
Exploring allosteric communication in multiple states of the bacterial ribosome using residue network analysis.Turk J Biol. 2018 Oct 25;42(5):392-404. doi: 10.3906/biy-1802-77. eCollection 2018. Turk J Biol. 2018. PMID: 30930623 Free PMC article.
-
Histidine methylation of yeast ribosomal protein Rpl3p is required for proper 60S subunit assembly.Mol Cell Biol. 2014 Aug;34(15):2903-16. doi: 10.1128/MCB.01634-13. Epub 2014 May 27. Mol Cell Biol. 2014. PMID: 24865971 Free PMC article.
-
Ribosomal protein uS19 mutants reveal its role in coordinating ribosome structure and function.Translation (Austin). 2015 Nov 18;3(2):e1117703. doi: 10.1080/21690731.2015.1117703. eCollection 2015 Jul-Dec. Translation (Austin). 2015. PMID: 26824029 Free PMC article.
-
Tracking fluctuation hotspots on the yeast ribosome through the elongation cycle.Nucleic Acids Res. 2017 May 5;45(8):4958-4971. doi: 10.1093/nar/gkx112. Nucleic Acids Res. 2017. PMID: 28334755 Free PMC article.
References
-
- Dunkle JA, Cate JH. Ribosome structure and dynamics during translocation and termination. Annu. Rev. Biophys. 2010;39:227–244. - PubMed
-
- Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem. 2001;70:415–435. - PubMed
-
- Aitken CE, Petrov A, Puglisi JD. Single ribosome dynamics and the mechanism of translation. Annu. Rev. Biophys. 2010;39:491–513. - PubMed
-
- Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

