Gene duplication and environmental adaptation within yeast populations
- PMID: 20660110
- PMCID: PMC2997561
- DOI: 10.1093/gbe/evq043
Gene duplication and environmental adaptation within yeast populations
Abstract
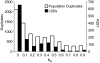
Population-level differences in the number of copies of genes resulting from gene duplication and loss have recently been recognized as an important source of variation in eukaryotes. However, except for a small number of cases, the phenotypic effects of this variation are unknown. Data from the Saccharomyces Genome Resequencing Project permit the study of duplication in genome sequences from a set of individuals within the same population. These sequences can be correlated with available information on the environments from which these yeast strains were isolated. We find that yeast show an abundance of duplicate genes that are lineage specific, leading to a large degree of variation in gene content between individual strains. There is a detectable bias for specific functions, indicating that selection is acting to preferentially retain certain duplicates. Most strikingly, we find that sets of over- and underrepresented duplicates correlate with the environment from which they were isolated. Together, these observations indicate that gene duplication can give rise to substantial phenotypic differences within populations that in turn can offer a shortcut to evolutionary adaptation.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

