Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts
- PMID: 20660200
- PMCID: PMC2937766
- DOI: 10.1128/JVI.00707-10
Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts
Abstract
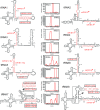
We applied deep sequencing technology to small RNA fractions from cells lytically infected with murine gammaherpesvirus 68 (gammaHV68) in order to define in detail small RNAs generated from a cluster of tRNA-related polycistronic structures located at the left end of the viral genome. We detected 10 new candidate microRNAs (miRNAs), six of which were confirmed by Northern blot analysis, leaving four as provisional. In addition, we determined that previously identified and annotated viral miRNA molecules were not necessarily represented as the most abundant sequence originating from a transcript. Based on these new small RNAs and previously reported gammaHV68 miRNAs, we were able to further describe and annotate the distinctive gammaHV68 tRNA-miRNA structures. We used this deep sequencing data and computational analysis to identify similar structures in the mouse genome and validated that these host structures also give rise to small RNAs. This reveals a possible convergent usage of tRNA/polymerase III (pol III) transcripts to generate small RNAs from both mammalian and viral genomes.
Figures


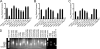


References
-
- Borchert, G. M., W. Lanier, and B. L. Davidson. 2006. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 13:1097-1101. - PubMed
-
- Bowden, R. J., J. P. Simas, A. J. Davis, and S. Efstathiou. 1997. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J. Gen. Virol. 78:1675-1687. - PubMed
-
- Cook, H. L., J. R. Lytle, H. E. Mischo, M. J. Li, J. J. Rossi, D. P. Silva, R. C. Desrosiers, and J. A. Steitz. 2005. Small nuclear RNAs encoded by herpesvirus saimiri upregulate the expression of genes linked to T cell activation in virally transformed T cells. Curr. Biol. 15:974-979. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

