KLF6 Gene and early melanoma development in a collagen I-rich extracellular environment
- PMID: 20660366
- PMCID: PMC2914760
- DOI: 10.1093/jnci/djq218
KLF6 Gene and early melanoma development in a collagen I-rich extracellular environment
Abstract
Background: A putative tumor suppressor gene at chromosome 10p15, which contains KLF6 and other genes, is predicted to be lost during melanoma development, and its identity is unknown. In this study, we investigated the biological roles and identity of this tumor suppressor gene.
Methods: The human UACC 903 melanoma cell line containing introduced DNA fragments from the 10p15 region with (10E6/3, 10E6/11, and 10E6/18) and without (10ER4S.2/1) the tumor suppressor gene was used. Xenograft tumors were generated in a total of 40 mice with melanoma cell lines, and tumor size was measured. Cells were cultured on plastic or a gel of type I collagen. Viability, proliferation, and apoptosis were assessed. Expression of KLF6 protein was assessed by immunohistochemistry and immunoblot analysis. Expression of phosphorylated Erk1/2 and cyclin D1 was assessed by immunoblot analysis. Protein expression of KLF6 was inhibited with small interfering RNA (siRNA). KLF6 protein expression was assessed in 17 human nevi and human melanoma specimens from 29 patients. Statistical analyses were adjusted for multiple comparisons by use of Dunnett method. All statistical tests were two-sided.
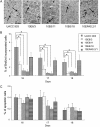
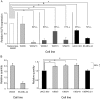
Results: Melanoma cells containing KLF6 generated smaller subcutaneous xenograft tumors with fewer proliferating cells than control cells. When grown on collagen 1, viability of cells with ectopic KLF6 expression (72%) was lower than that of control cells (100%) (group difference = -28%, 95% confidence interval = -31.3% to -25.2%, P < .001). Viability of melanoma cells with or without the KLF6 tumor suppressor gene on plastic dishes was similar. When KLF6 expression was inhibited with KLF6 siRNA, viability of cells with the tumor suppressor gene on collagen I gel increased compared with that of control cells carrying scrambled siRNA. KLF6 protein was detected in all nevi examined but not in human metastatic melanoma tissue examined. Ectopic expression of KLF6 protein in melanoma cells grown on collagen I decreased levels of phosphorylated Erk1/2 and cyclin D1 in the mitogen-activated protein kinase signaling pathway.
Conclusions: In melanoma cells, the tumor suppressor gene at 10p15 appears to be KLF6. Signaling from the collagen I-rich extracellular matrix appears to be involved in the tumor suppressive activity of KLF6 protein.
Figures






Comment in
-
The ups and downs of transcription factors in melanoma.J Natl Cancer Inst. 2010 Aug 4;102(15):1103-4. doi: 10.1093/jnci/djq267. Epub 2010 Jul 21. J Natl Cancer Inst. 2010. PMID: 20660367 No abstract available.
References
-
- Newton JA. Genetics of melanoma. Br Med Bull. 1994;50(3):677–687. - PubMed
-
- Robertson GP, Herbst RA, Nagane M, Huang HJ, Cavenee WK. The chromosome 10 monosomy common in human melanomas results from loss of two separate tumor suppressor loci. Cancer Res. 1999;59(15):3596–3601. - PubMed
-
- Isshiki K, Elder DE, Guerry D, Linnenbach AJ. Chromosome 10 allelic loss in malignant melanoma. Genes Chromosomes Cancer. 1993;8(3):178–184. - PubMed
-
- Bastian BC, LeBoit PE, Hamm H, Brocker EB, Pinkel D. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998;58(10):2170–2175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

