Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila
- PMID: 20660614
- PMCID: PMC2916131
- DOI: 10.1084/jem.20100771
Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila
Abstract
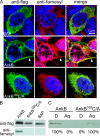
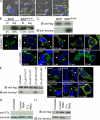
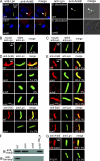
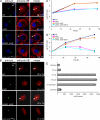
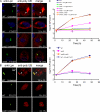
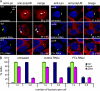
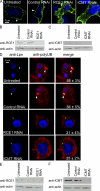
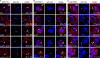
Farnesylation involves covalent linkage of eukaryotic proteins to a lipid moiety to anchor them into membranes, which is essential for the biological function of Ras and other proteins. A large cadre of bacterial effectors is injected into host cells by intravacuolar pathogens through elaborate type III-VII translocation machineries, and many of these effectors are incorporated into the pathogen-containing vacuolar membrane by unknown mechanisms. The Dot/Icm type IV secretion system of Legionella pneumophila injects into host cells the F-box effector Ankyrin B (AnkB), which functions as platforms for the docking of polyubiquitinated proteins to the Legionella-containing vacuole (LCV) to enable intravacuolar proliferation in macrophages and amoeba. We show that farnesylation of AnkB is indispensable for its anchoring to the cytosolic face of the LCV membrane, for its biological function within macrophages and Dictyostelium discoideum, and for intrapulmonary proliferation in mice. Remarkably, the protein farnesyltransferase, RCE-1 (Ras-converting enzyme-1), and isoprenyl cysteine carboxyl methyltransferase host farnesylation enzymes are recruited to the LCV in a Dot/Icm-dependent manner and are essential for the biological function of AnkB. In conclusion, this study shows novel localized recruitment of the host farnesylation machinery and its anchoring of an F-box effector to the LCV membrane, and this is essential for biological function in vitro and in vivo.
Figures