Targeting DNA double-strand breaks with TAL effector nucleases
- PMID: 20660643
- PMCID: PMC2942870
- DOI: 10.1534/genetics.110.120717
Targeting DNA double-strand breaks with TAL effector nucleases
Abstract
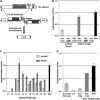
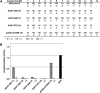
Engineered nucleases that cleave specific DNA sequences in vivo are valuable reagents for targeted mutagenesis. Here we report a new class of sequence-specific nucleases created by fusing transcription activator-like effectors (TALEs) to the catalytic domain of the FokI endonuclease. Both native and custom TALE-nuclease fusions direct DNA double-strand breaks to specific, targeted sites.
Figures
References
-
- Boch, J., H. Scholze, S. Schornack, A. Landgraf, S. Hahn et al., 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326 1509–1512. - PubMed
-
- Bogdanove, A. J., S. Schornack and T. Lahaye, 2010. TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13 394–401. - PubMed
-
- Bonas, U., R. E. Stall and B. Staskawicz, 1989. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. 218 127–136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

