Identification of a third variable lymphocyte receptor in the lamprey
- PMID: 20660745
- PMCID: PMC2922563
- DOI: 10.1073/pnas.1001910107
Identification of a third variable lymphocyte receptor in the lamprey
Abstract
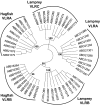
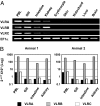
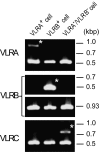
Jawless vertebrates such as lamprey and hagfish lack T-cell and B-cell receptors; instead, they have unique antigen receptors known as variable lymphocyte receptors (VLRs). VLRs generate diversity by recombining highly diverse leucine-rich repeat modules and are expressed clonally on lymphocyte-like cells (LLCs). Thus far, two types of receptors, VLRA and VLRB, have been identified in lampreys and hagfish. Recent evidence indicates that VLRA and VLRB are expressed on distinct populations of LLCs that resemble T cells and B cells of jawed vertebrates, respectively. Here we identified a third VLR, designated VLRC, in the lamprey. None of the approximately 100 VLRC cDNA clones subjected to sequencing had an identical sequence, indicating that VLRC can generate sufficient diversity to function as antigen receptors. Notably, the C-terminal cap of VLRC exhibits only limited diversity and has important structural differences relative to VLRA and VLRB. Single-cell PCR analysis identified LLCs that rearranged VLRC but not VLRA or VLRB, suggesting the presence of a unique population of LLCs that express only VLRC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Boffa GA, Fine JM, Drilhon A, Amouch P. Immunoglobulins and transferrin in marine lamprey sera. Nature. 1967;214:700–702. - PubMed
-
- Linthicum DS, Hildemann WH. Immunologic responses of Pacific hagfish. 3. Serum antibodies to cellular antigens. J Immunol. 1970;105:912–918. - PubMed
-
- Pollara B, Litman GW, Finstad J, Howell J, Good RA. The evolution of the immune response. VII. Antibody to human “O” cells and properties of the immunoglobulin in lamprey. J Immunol. 1970;105:738–745. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

