Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families
- PMID: 20661309
- PMCID: PMC2908706
- DOI: 10.1371/journal.pgen.1001036
Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families
Abstract
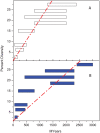
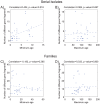
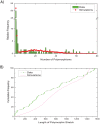
Our understanding of basic evolutionary processes in bacteria is still very limited. For example, multiple recent dating estimates are based on a universal inter-species molecular clock rate, but that rate was calibrated using estimates of geological dates that are no longer accepted. We therefore estimated the short-term rates of mutation and recombination in Helicobacter pylori by sequencing an average of 39,300 bp in 78 gene fragments from 97 isolates. These isolates included 34 pairs of sequential samples, which were sampled at intervals of 0.25 to 10.2 years. They also included single isolates from 29 individuals (average age: 45 years) from 10 families. The accumulation of sequence diversity increased with time of separation in a clock-like manner in the sequential isolates. We used Approximate Bayesian Computation to estimate the rates of mutation, recombination, mean length of recombination tracts, and average diversity in those tracts. The estimates indicate that the short-term mutation rate is 1.4 x 10(-6) (serial isolates) to 4.5 x 10(-6) (family isolates) per nucleotide per year and that three times as many substitutions are introduced by recombination as by mutation. The long-term mutation rate over millennia is 5-17-fold lower, partly due to the removal of non-synonymous mutations due to purifying selection. Comparisons with the recent literature show that short-term mutation rates vary dramatically in different bacterial species and can span a range of several orders of magnitude.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

