Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast
- PMID: 20661445
- PMCID: PMC2908685
- DOI: 10.1371/journal.pgen.1001032
Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast
Abstract
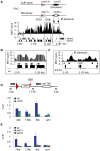
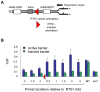
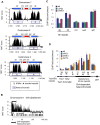
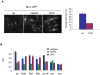
Schizosaccharomyces pombe Rad3 checkpoint kinase and its human ortholog ATR are essential for maintaining genome integrity in cells treated with genotoxins that damage DNA or arrest replication forks. Rad3 and ATR also function during unperturbed growth, although the events triggering their activation and their critical functions are largely unknown. Here, we use ChIP-on-chip analysis to map genomic loci decorated by phosphorylated histone H2A (gammaH2A), a Rad3 substrate that establishes a chromatin-based recruitment platform for Crb2 and Brc1 DNA repair/checkpoint proteins. Unexpectedly, gammaH2A marks a diverse array of genomic features during S-phase, including natural replication fork barriers and a fork breakage site, retrotransposons, heterochromatin in the centromeres and telomeres, and ribosomal RNA (rDNA) repeats. gammaH2A formation at the centromeres and telomeres is associated with heterochromatin establishment by Clr4 histone methyltransferase. We show that gammaH2A domains recruit Brc1, a factor involved in repair of damaged replication forks. Brc1 C-terminal BRCT domain binding to gammaH2A is crucial in the absence of Rqh1(Sgs1), a RecQ DNA helicase required for rDNA maintenance whose human homologs are mutated in patients with Werner, Bloom, and Rothmund-Thomson syndromes that are characterized by cancer-predisposition or accelerated aging. We conclude that Rad3 phosphorylates histone H2A to mobilize Brc1 to critical genomic domains during S-phase, and this pathway functions in parallel with Rqh1 DNA helicase in maintaining genome integrity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. - PubMed
-
- Boddy MN, Russell P. DNA replication checkpoint. Curr Biol. 2001;11:R953–956. - PubMed
-
- Pearson C, Nichol Edamura K, Cleary J. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

