The Caenorhabditis elegans homolog of Gen1/Yen1 resolvases links DNA damage signaling to DNA double-strand break repair
- PMID: 20661466
- PMCID: PMC2908289
- DOI: 10.1371/journal.pgen.1001025
The Caenorhabditis elegans homolog of Gen1/Yen1 resolvases links DNA damage signaling to DNA double-strand break repair
Abstract
DNA double-strand breaks (DSBs) can be repaired by homologous recombination (HR), which can involve Holliday junction (HJ) intermediates that are ultimately resolved by nucleolytic enzymes. An N-terminal fragment of human GEN1 has recently been shown to act as a Holliday junction resolvase, but little is known about the role of GEN-1 in vivo. Holliday junction resolution signifies the completion of DNA repair, a step that may be coupled to signaling proteins that regulate cell cycle progression in response to DNA damage. Using forward genetic approaches, we identified a Caenorhabditis elegans dual function DNA double-strand break repair and DNA damage signaling protein orthologous to the human GEN1 Holliday junction resolving enzyme. GEN-1 has biochemical activities related to the human enzyme and facilitates repair of DNA double-strand breaks, but is not essential for DNA double-strand break repair during meiotic recombination. Mutational analysis reveals that the DNA damage-signaling function of GEN-1 is separable from its role in DNA repair. GEN-1 promotes germ cell cycle arrest and apoptosis via a pathway that acts in parallel to the canonical DNA damage response pathway mediated by RPA loading, CHK1 activation, and CEP-1/p53-mediated apoptosis induction. Furthermore, GEN-1 acts redundantly with the 9-1-1 complex to ensure genome stability. Our study suggests that GEN-1 might act as a dual function Holliday junction resolvase that may coordinate DNA damage signaling with a late step in DNA double-strand break repair.
Conflict of interest statement
The authors have declared that no competing interests exist.
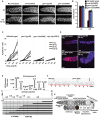
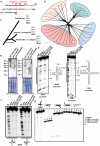
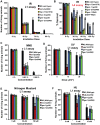
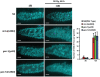
Figures
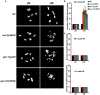
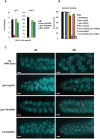


References
-
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. - PubMed
-
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. - PubMed
-
- Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 2000;10:39–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

