Determining gene flow and the influence of selection across the equatorial barrier of the East Pacific Rise in the tube-dwelling polychaete Alvinella pompejana
- PMID: 20663123
- PMCID: PMC2924869
- DOI: 10.1186/1471-2148-10-220
Determining gene flow and the influence of selection across the equatorial barrier of the East Pacific Rise in the tube-dwelling polychaete Alvinella pompejana
Abstract
Background: Comparative phylogeography recently performed on the mitochondrial cytochrome oxidase I (mtCOI) gene from seven deep-sea vent species suggested that the East Pacific Rise fauna has undergone a vicariant event with the emergence of a north/south physical barrier at the Equator 1-2 Mya. Within this specialised fauna, the tube-dwelling polychaete Alvinella pompejana showed reciprocal monophyly at mtCOI on each side of the Equator (9 degrees 50'N/7 degrees 25'S), suggesting potential, ongoing allopatric speciation. However, the development of a barrier to gene flow is a long and complex process. Secondary contact between previously isolated populations can occur when physical isolation has not persisted long enough to result in reproductive isolation between genetically divergent lineages, potentially leading to hybridisation and subsequent allelic introgression. The present study evaluates the strength of the equatorial barrier to gene flow and tests for potential secondary contact zones between A. pompejana populations by comparing the mtCOI gene with nuclear genes.
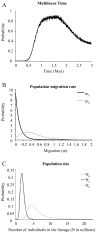
Results: Allozyme frequencies and the analysis of nucleotide polymorphisms at three nuclear loci confirmed the north/south genetic differentiation of Alvinella pompejana populations along the East Pacific Rise. Migration was oriented north-to-south with a moderate allelic introgression between the two geographic groups over a narrow geographic range just south of the barrier. Multilocus analysis also indicated that southern populations have undergone demographic expansion as previously suggested by a multispecies approach. A strong shift in allozyme frequencies together with a high level of divergence between alleles and a low number of 'hybrid' individuals were observed between the northern and southern groups using the phosphoglucomutase gene. In contrast, the S-adenosylhomocysteine hydrolase gene exhibited reduced diversity and a lack of population differentiation possibly due to a selective sweep or hitch-hiking.
Conclusions: The equatorial barrier leading to the separation of East Pacific Rise vent fauna into two distinct geographic groups is still permeable to migration, with a probable north-to-south migration route for A. pompejana. This separation also coincides with demographic expansion in the southern East Pacific Rise. Our results suggest that allopatry resulting from ridge offsetting is a common mechanism of speciation for deep-sea hydrothermal vent organisms.
Figures



Similar articles
-
Population subdivision of hydrothermal vent polychaete Alvinella pompejana across equatorial and Easter Microplate boundaries.BMC Evol Biol. 2016 Oct 28;16(1):235. doi: 10.1186/s12862-016-0807-9. BMC Evol Biol. 2016. PMID: 27793079 Free PMC article.
-
Comparative phylogeography among hydrothermal vent species along the East Pacific Rise reveals vicariant processes and population expansion in the South.Mol Ecol. 2009 Sep;18(18):3903-17. doi: 10.1111/j.1365-294X.2009.04325.x. Epub 2009 Aug 26. Mol Ecol. 2009. PMID: 19709370
-
Genomic patterns of divergence in the early and late steps of speciation of the deep-sea vent thermophilic worms of the genus Alvinella.BMC Ecol Evol. 2022 Sep 3;22(1):106. doi: 10.1186/s12862-022-02057-y. BMC Ecol Evol. 2022. PMID: 36057769 Free PMC article.
-
Thermal selection of PGM allozymes in newly founded populations of the thermotolerant vent polychaete Alvinella pompejana.Proc Biol Sci. 2004 Nov 22;271(1555):2351-9. doi: 10.1098/rspb.2004.2852. Proc Biol Sci. 2004. PMID: 15556887 Free PMC article.
-
Does selection favour the maintenance of porous species boundaries?J Evol Biol. 2024 Jun 28;37(6):616-627. doi: 10.1093/jeb/voae030. J Evol Biol. 2024. PMID: 38599591 Review.
Cited by
-
Genetic connectivity between north and south Mid-Atlantic Ridge chemosynthetic bivalves and their symbionts.PLoS One. 2012;7(7):e39994. doi: 10.1371/journal.pone.0039994. Epub 2012 Jul 6. PLoS One. 2012. PMID: 22792208 Free PMC article.
-
262 Voyages Beneath the Sea: a global assessment of macro- and megafaunal biodiversity and research effort at deep-sea hydrothermal vents.PeerJ. 2019 Aug 6;7:e7397. doi: 10.7717/peerj.7397. eCollection 2019. PeerJ. 2019. PMID: 31404427 Free PMC article.
-
Antagonistic evolution of an antibiotic and its molecular chaperone: how to maintain a vital ectosymbiosis in a highly fluctuating habitat.Sci Rep. 2017 May 3;7(1):1454. doi: 10.1038/s41598-017-01626-2. Sci Rep. 2017. PMID: 28469247 Free PMC article.
-
Population structure of Bathymodiolus manusensis, a deep-sea hydrothermal vent-dependent mussel from Manus Basin, Papua New Guinea.PeerJ. 2017 Aug 21;5:e3655. doi: 10.7717/peerj.3655. eCollection 2017. PeerJ. 2017. PMID: 28852590 Free PMC article.
-
Balanced Polymorphism at the Pgm-1 Locus of the Pompeii Worm Alvinella pompejana and Its Variant Adaptability Is Only Governed by Two QE Mutations at Linked Sites.Genes (Basel). 2022 Jan 24;13(2):206. doi: 10.3390/genes13020206. Genes (Basel). 2022. PMID: 35205251 Free PMC article.
References
-
- Malécot G. Quelques schémas probabilistes sur la variabilité des populations naturelles. Ann Univ Lyon Sci. 1950;13:37–60.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

