X-ray diffraction experiment--the last experiment in the structure elucidation process
- PMID: 20663480
- PMCID: PMC3128764
- DOI: 10.1016/S1876-1623(09)77002-6
X-ray diffraction experiment--the last experiment in the structure elucidation process
Abstract
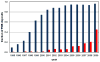
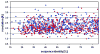
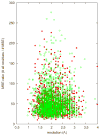
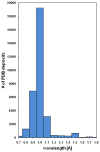
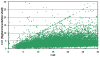
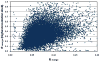
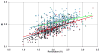
Recent years have brought not only an avalanche of new macromolecular structures, but also significant advances in the protein structure determination methodology only now making its way into structure-based drug discovery. In this chapter, we review recent methodology developments in X-ray diffraction experiments that led to fast and very accurate elucidation of three-dimensional structures of macromolecules. We will discuss the role of data collection as the last experiment performed in the crystal structure determination process. A statistical analysis of diffraction experiments that are reported in the Protein Data Bank (PDB) is also presented.
Copyright 2009 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
X-ray crystallography as a tool for mechanism-of-action studies and drug discovery.Curr Pharm Biotechnol. 2013;14(5):537-50. doi: 10.2174/138920101405131111104824. Curr Pharm Biotechnol. 2013. PMID: 22429136 Review.
-
The future of crystallography in drug discovery.Expert Opin Drug Discov. 2014 Feb;9(2):125-37. doi: 10.1517/17460441.2014.872623. Epub 2013 Dec 28. Expert Opin Drug Discov. 2014. PMID: 24372145 Free PMC article. Review.
-
A public database of macromolecular diffraction experiments.Acta Crystallogr D Struct Biol. 2016 Nov 1;72(Pt 11):1181-1193. doi: 10.1107/S2059798316014716. Epub 2016 Oct 28. Acta Crystallogr D Struct Biol. 2016. PMID: 27841751 Free PMC article.
-
Protein crystallization.Methods Mol Biol. 2007;383:337-49. doi: 10.1007/978-1-59745-335-6_22. Methods Mol Biol. 2007. PMID: 18217696
-
X-ray sources and high-throughput data collection methods.Methods Mol Biol. 2012;841:93-141. doi: 10.1007/978-1-61779-520-6_5. Methods Mol Biol. 2012. PMID: 22222450
Cited by
-
Macrocyclic Tetramers-Structural Investigation of Peptide-Peptoid Hybrids.Molecules. 2021 Jul 28;26(15):4548. doi: 10.3390/molecules26154548. Molecules. 2021. PMID: 34361700 Free PMC article.
-
Recent Force Field Strategies for Intrinsically Disordered Proteins.J Chem Inf Model. 2021 Mar 22;61(3):1037-1047. doi: 10.1021/acs.jcim.0c01175. Epub 2021 Feb 16. J Chem Inf Model. 2021. PMID: 33591749 Free PMC article. Review.
-
Unmet challenges of structural genomics.Curr Opin Struct Biol. 2010 Oct;20(5):587-97. doi: 10.1016/j.sbi.2010.08.001. Epub 2010 Aug 31. Curr Opin Struct Biol. 2010. PMID: 20810277 Free PMC article. Review.
-
X-ray crystallography: Assessment and validation of protein-small molecule complexes for drug discovery.Expert Opin Drug Discov. 2011 Aug 1;6(8):771-782. doi: 10.1517/17460441.2011.585154. Expert Opin Drug Discov. 2011. PMID: 21779303 Free PMC article.
-
100 Years later: Celebrating the contributions of x-ray crystallography to allergy and clinical immunology.J Allergy Clin Immunol. 2015 Jul;136(1):29-37.e10. doi: 10.1016/j.jaci.2015.05.016. J Allergy Clin Immunol. 2015. PMID: 26145985 Free PMC article. Review.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Crystallogr. 2002;D58:1948–1954. - PubMed
-
- Ascone I, Strange R. J Synchrotron Radiat. 2009;16:413–421. - PubMed
-
- Bech M, Bunk O, David C, Kraft P, Bronnimann C, Eikenberry EF, Pfeiffer F. Appl Radiat Isot. 2008;66:474–478. - PubMed
-
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. Acta Crystallogr. 2002;D58:899–907. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

