Structure-guided design and immunological characterization of immunogens presenting the HIV-1 gp120 V3 loop on a CTB scaffold
- PMID: 20663531
- PMCID: PMC2942080
- DOI: 10.1016/j.virol.2010.06.027
Structure-guided design and immunological characterization of immunogens presenting the HIV-1 gp120 V3 loop on a CTB scaffold
Erratum in
- Virology. 2011 Jan 20;409(2):360. Abagyan, Ruben [added]
Abstract
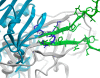
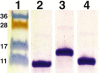
V3 loop is a major neutralizing determinant of the HIV-1 gp120. Using 3D structures of cholera toxin B subunit (CTB), complete V3 in the gp120 context, and V3 bound to a monoclonal antibody (mAb), we designed two V3-scaffold immunogen constructs (V3-CTB). The full-length V3-CTB presenting the complete V3 in a structural context mimicking gp120 was recognized by the large majority of our panel of 24 mAbs. The short V3-CTB presenting a V3 fragment in the conformation observed in the complex with the 447-52D Fab, exhibited high-affinity binding to this mAb. The immunogens were evaluated in rabbits using DNA-prime/protein-boost protocol. Boosting with the full-length V3-CTB induced high anti-V3 titers in sera that potently neutralize multiple HIV virus strains. The short V3-CTB was ineffective. The results suggest that very narrow antigenic profile of an immunogen is associated with poor Ab response. An immunogen with broader antigenic activity elicits robust Ab response.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures




References
-
-
Immune epitope Database and Analysis Resource. In.
-
-
- Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983–1002. - PubMed
-
- Abagyan R, Totrov M, Kuznetsov D. ICM-A new method for protein modeling and design: Applications to. J Comp Chem. 1994;15:488–506.
-
- Backstrom M, Lebens M, Schodel F, Holmgren J. Insertion of a HIV-1-neutralizing epitope in a surface-exposed internal region of the cholera toxin B-subunit. Gene. 1994;149:211–217. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

