Characterization and prediction of protein nucleolar localization sequences
- PMID: 20663773
- PMCID: PMC2995072
- DOI: 10.1093/nar/gkq653
Characterization and prediction of protein nucleolar localization sequences
Abstract
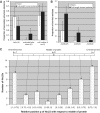
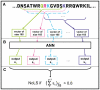
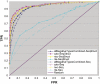
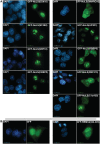
Although the nucleolar localization of proteins is often believed to be mediated primarily by non-specific retention to core nucleolar components, many examples of short nucleolar targeting sequences have been reported in recent years. In this article, 46 human nucleolar localization sequences (NoLSs) were collated from the literature and subjected to statistical analysis. Of the residues in these NoLSs 48% are basic, whereas 99% of the residues are predicted to be solvent-accessible with 42% in α-helix and 57% in coil. The sequence and predicted protein secondary structure of the 46 NoLSs were used to train an artificial neural network to identify NoLSs. At a true positive rate of 54%, the predictor's overall false positive rate (FPR) is estimated to be 1.52%, which can be broken down to FPRs of 0.26% for randomly chosen cytoplasmic sequences, 0.80% for randomly chosen nucleoplasmic sequences and 12% for nuclear localization signals. The predictor was used to predict NoLSs in the complete human proteome and 10 of the highest scoring previously unknown NoLSs were experimentally confirmed. NoLSs are a prevalent type of targeting motif that is distinct from nuclear localization signals and that can be computationally predicted.
Figures
References
-
- Scheer U, Hock R. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 1999;11:385–390. - PubMed
-
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. - PubMed
-
- Olson MO, Dundr M, Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

