Apical scaffolding protein NHERF2 modulates the localization of alternatively spliced plasma membrane Ca2+ pump 2B variants in polarized epithelial cells
- PMID: 20663896
- PMCID: PMC2951242
- DOI: 10.1074/jbc.M110.164137
Apical scaffolding protein NHERF2 modulates the localization of alternatively spliced plasma membrane Ca2+ pump 2B variants in polarized epithelial cells
Abstract
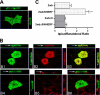
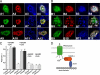
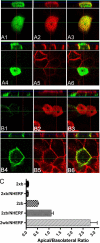
The membrane localization of the plasma membrane Ca(2+)-ATPase isoform 2 (PMCA2) in polarized cells is determined by alternative splicing; the PMCA2w/b splice variant shows apical localization, whereas the PMCA2z/b and PMCA2x/b variants are mostly basolateral. We previously reported that PMCA2b interacts with the PDZ protein Na(+)/H(+) exchanger regulatory factor 2 (NHERF2), but the role of this interaction for the specific membrane localization of PMCA2 is not known. Here we show that co-expression of NHERF2 greatly enhanced the apical localization of GFP-tagged PMCA2w/b in polarized Madin-Darby canine kidney cells. GFP-PMCA2z/b was also redirected to the apical membrane by NHERF2, whereas GFP-PMCA2x/b remained exclusively basolateral. In the presence of NHERF2, GFP-PMCA2w/b co-localized with the actin-binding protein ezrin even after disruption of the actin cytoskeleton by cytochalasin D or latrunculin B. Surface biotinylation and fluorescence recovery after photobleaching experiments demonstrated that NHERF2-mediated anchorage to the actin cytoskeleton reduced internalization and lateral mobility of the pump. Our results show that the specific interaction with NHERF2 enhances the apical concentration of PMCA2w/b by anchoring the pump to the apical membrane cytoskeleton. The data also suggest that the x/b splice form of PMCA2 contains a dominant lateral targeting signal, whereas the targeting and localization of the z/b form are more flexible and not fully determined by intrinsic sequence features.
Figures

Similar articles
-
Apical localization of PMCA2w/b is enhanced in terminally polarized MDCK cells.Biochem Biophys Res Commun. 2011 Jul 1;410(2):322-7. doi: 10.1016/j.bbrc.2011.05.147. Epub 2011 Jun 6. Biochem Biophys Res Commun. 2011. PMID: 21672522 Free PMC article.
-
Regulation of apical membrane enrichment and retention of plasma membrane Ca ATPase splice variants by the PDZ-domain protein NHERF2.Commun Integr Biol. 2011 May;4(3):340-3. doi: 10.4161/cib.4.3.15040. Epub 2011 May 1. Commun Integr Biol. 2011. PMID: 21980575 Free PMC article.
-
Plasma membrane Ca2+ ATPase isoform 2b interacts preferentially with Na+/H+ exchanger regulatory factor 2 in apical plasma membranes.J Biol Chem. 2002 Mar 22;277(12):10506-11. doi: 10.1074/jbc.M111616200. Epub 2002 Jan 10. J Biol Chem. 2002. PMID: 11786550
-
CFTR-NHERF2-LPA₂ Complex in the Airway and Gut Epithelia.Int J Mol Sci. 2017 Sep 4;18(9):1896. doi: 10.3390/ijms18091896. Int J Mol Sci. 2017. PMID: 28869532 Free PMC article. Review.
-
The epithelial brush border Na+/H+ exchanger NHE3 associates with the actin cytoskeleton by binding to ezrin directly and via PDZ domain-containing Na+/H+ exchanger regulatory factor (NHERF) proteins.Clin Exp Pharmacol Physiol. 2008 Aug;35(8):863-71. doi: 10.1111/j.1440-1681.2008.04931.x. Epub 2008 Apr 21. Clin Exp Pharmacol Physiol. 2008. PMID: 18430067 Review.
Cited by
-
Differential expression of PMCA2 mRNA isoforms in a cohort of Spanish patients with breast tumor types.Oncol Lett. 2018 Dec;16(6):6950-6959. doi: 10.3892/ol.2018.9540. Epub 2018 Oct 2. Oncol Lett. 2018. PMID: 30546427 Free PMC article.
-
Activated ERM protein plays a critical role in drug resistance of MOLT4 cells induced by CCL25.PLoS One. 2013;8(1):e52384. doi: 10.1371/journal.pone.0052384. Epub 2013 Jan 9. PLoS One. 2013. PMID: 23326330 Free PMC article.
-
NHERF1 Is Required for Localization of PMCA2 and Suppression of Early Involution in the Female Lactating Mammary Gland.Endocrinology. 2019 Aug 1;160(8):1797-1810. doi: 10.1210/en.2019-00230. Endocrinology. 2019. PMID: 31087002 Free PMC article.
-
Emanuel Strehler's work on calcium pumps and calcium signaling.World J Biol Chem. 2011 Apr 26;2(4):67-72. doi: 10.4331/wjbc.v2.i4.67. World J Biol Chem. 2011. PMID: 21537475 Free PMC article.
-
PMCA2 regulates HER2 protein kinase localization and signaling and promotes HER2-mediated breast cancer.Proc Natl Acad Sci U S A. 2016 Jan 19;113(3):E282-90. doi: 10.1073/pnas.1516138113. Epub 2016 Jan 4. Proc Natl Acad Sci U S A. 2016. PMID: 26729871 Free PMC article.
References
-
- Strehler E. E., Zacharias D. A. (2001) Physiol. Rev. 81, 21–50 - PubMed
-
- Chicka M. C., Strehler E. E. (2003) J. Biol. Chem. 278, 18464–18470 - PubMed
-
- Grati M., Aggarwal N., Strehler E. E., Wenthold R. J. (2006) J. Cell Sci. 119, 2995–3007 - PubMed
-
- DeMarco S. J., Strehler E. E. (2001) J. Biol. Chem. 276, 21594–21600 - PubMed
-
- Schuh K., Uldrijan S., Gambaryan S., Roethlein N., Neyses L. (2003) J. Biol. Chem. 278, 9778–9883 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

