Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer
- PMID: 20663909
- PMCID: PMC2922412
- DOI: 10.1158/0008-5472.CAN-10-0582
Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer
Abstract
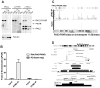
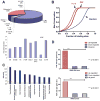
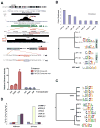
The PAX3-FKHR fusion protein is present in a majority of alveolar rhabdomyosarcomas associated with increased aggressiveness and poor prognosis. To better understand the molecular pathogenesis of PAX3-FKHR, we carried out the first, unbiased genome-wide identification of PAX3-FKHR binding sites and associated target genes in alveolar rhabdomyosarcoma. The data shows that PAX3-FKHR binds to the same sites as PAX3 at both MYF5 and MYOD enhancers. The genome-wide analysis reveals that the PAX3-FKHR sites are (a) mostly distal to transcription start sites, (b) conserved, (c) enriched for PAX3 motifs, and (d) strongly associated with genes overexpressed in PAX3-FKHR-positive rhabdomyosarcoma cells and tumors. There is little evidence in our data set for PAX3-FKHR binding at the promoter sequences. The genome-wide analysis further illustrates a strong association between PAX3 and E-box motifs in these binding sites, suggestive of a common coregulation for many target genes. We also provide the first direct evidence that FGFR4 and IGF1R are the targets for PAX3-FKHR. The map of PAX3-FKHR binding sites provides a framework for understanding the pathogenic roles of PAX3-FKHR, as well as its molecular targets to allow a systematic evaluation of agents against this aggressive rhabdomyosarcoma.
(c)2010 AACR.
Figures


Similar articles
-
Carnitine palmitoyltransferase 1A (CPT1A): a transcriptional target of PAX3-FKHR and mediates PAX3-FKHR-dependent motility in alveolar rhabdomyosarcoma cells.BMC Cancer. 2012 Apr 25;12:154. doi: 10.1186/1471-2407-12-154. BMC Cancer. 2012. PMID: 22533991 Free PMC article.
-
Identification of PAX3-FKHR-regulated genes differentially expressed between alveolar and embryonal rhabdomyosarcoma: focus on MYCN as a biologically relevant target.Genes Chromosomes Cancer. 2008 Jun;47(6):510-20. doi: 10.1002/gcc.20554. Genes Chromosomes Cancer. 2008. PMID: 18335505
-
PAX-FKHR function as pangenes by simultaneously inducing and inhibiting myogenesis.Oncogene. 2008 Mar 27;27(14):2004-14. doi: 10.1038/sj.onc.1210835. Epub 2007 Oct 8. Oncogene. 2008. PMID: 17922034
-
Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma.Oncogene. 2001 Sep 10;20(40):5736-46. doi: 10.1038/sj.onc.1204599. Oncogene. 2001. PMID: 11607823 Review.
-
Alveolar rhabdomyosarcoma: is the cell of origin a mesenchymal stem cell?Cancer Lett. 2009 Jul 8;279(2):126-36. doi: 10.1016/j.canlet.2008.09.039. Epub 2008 Nov 12. Cancer Lett. 2009. PMID: 19008039 Review.
Cited by
-
Zebrafish rhabdomyosarcoma reflects the developmental stage of oncogene expression during myogenesis.Development. 2013 Jul;140(14):3040-50. doi: 10.1242/dev.087858. Development. 2013. PMID: 23821038 Free PMC article.
-
Differentiation-dependent chromosomal organization changes in normal myogenic cells are absent in rhabdomyosarcoma cells.Front Cell Dev Biol. 2023 Nov 7;11:1293891. doi: 10.3389/fcell.2023.1293891. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38020905 Free PMC article.
-
Understanding the Interplay between Expression, Mutation and Activity of ALK Receptor in Rhabdomyosarcoma Cells for Clinical Application of Small-Molecule Inhibitors.PLoS One. 2015 Jul 6;10(7):e0132330. doi: 10.1371/journal.pone.0132330. eCollection 2015. PLoS One. 2015. PMID: 26147305 Free PMC article.
-
Cell-based small-molecule compound screen identifies fenretinide as potential therapeutic for translocation-positive rhabdomyosarcoma.PLoS One. 2013;8(1):e55072. doi: 10.1371/journal.pone.0055072. Epub 2013 Jan 25. PLoS One. 2013. PMID: 23372815 Free PMC article.
-
SNAIL is a key regulator of alveolar rhabdomyosarcoma tumor growth and differentiation through repression of MYF5 and MYOD function.Cell Death Dis. 2018 May 29;9(6):643. doi: 10.1038/s41419-018-0693-8. Cell Death Dis. 2018. PMID: 29844345 Free PMC article.
References
-
- Blake JA, Thomas M, Thompson JA, White R, Ziman M. Perplexing Pax: from puzzle to paradigm. Dev Dyn. 2008;23710:2791–803. - PubMed
-
- Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–73. - PubMed
-
- Robson EJ, He SJ, Eccles MR. A PANorama of PAX genes in cancer and development. Nat Rev Cancer. 2006;61:52–62. - PubMed
-
- Lang D, Powell SK, Plummer RS, Young KP, Ruggeri BA. PAX genes: roles in development, pathophysiology, and cancer. Biochem Pharmacol. 2007;731:1–14. - PubMed
-
- Buckingham M. Skeletal muscle progenitor cells and the role of Pax genes. C R Biol. 2007;3306–7:530–3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

