Inhibition of transglutaminase 2 mitigates transcriptional dysregulation in models of Huntington disease
- PMID: 20665636
- PMCID: PMC3068019
- DOI: 10.1002/emmm.201000084
Inhibition of transglutaminase 2 mitigates transcriptional dysregulation in models of Huntington disease
Abstract
Caused by a polyglutamine expansion in the huntingtin protein, Huntington's disease leads to striatal degeneration via the transcriptional dysregulation of a number of genes, including those involved in mitochondrial biogenesis. Here we show that transglutaminase 2, which is upregulated in HD, exacerbates transcriptional dysregulation by acting as a selective corepressor of nuclear genes; transglutaminase 2 interacts directly with histone H3 in the nucleus. In a cellular model of HD, transglutaminase inhibition de-repressed two established regulators of mitochondrial function, PGC-1alpha and cytochrome c and reversed susceptibility of human HD cells to the mitochondrial toxin, 3-nitroproprionic acid; however, protection mediated by transglutaminase inhibition was not associated with improved mitochondrial bioenergetics. A gene microarray analysis indicated that transglutaminase inhibition normalized expression of not only mitochondrial genes but also 40% of genes that are dysregulated in HD striatal neurons, including chaperone and histone genes. Moreover, transglutaminase inhibition attenuated degeneration in a Drosophila model of HD and protected mouse HD striatal neurons from excitotoxicity. Altogether these findings demonstrate that selective TG inhibition broadly corrects transcriptional dysregulation in HD and defines a novel HDAC-independent epigenetic strategy for treating neurodegeneration.
Figures

Basal levels of PGC-1α mRNA are lower in immortalized striatal cells bearing mhtt with 111 CAG repeats (Q111) than in striatal cells bearing WT htt (Q7 cells) in accordance with prior studies. ZDON treatment increases PGC-1α mRNA levels in both Q7 and Q111 cells, while the catalytically inactive Zc has no effect.
Following the reduction in the expression of coactivator, PGC-1α, the nuclear encoded, mitochondrial gene, cytochrome c is diminished in Q111 cells. ZDON treatment of Q111 cells restores cytochrome c mRNA levels.
Three unique siRNA sequences targeted against TG2 (sequences 1–3) augmented cytochrome c and PGC-1α mRNA levels in Q111 cells at 24–48 h post-transfection relative to no transfection (No Trans) or scrambled siRNA (Control siRNA). The qualitatively similar result between distinct treatment groups indicates that the effect is target specific and not sequence specific. Each siRNA sequence reduced TG2 mRNA levels by greater than 70% (data not shown).
As expected from our ZDON and siRNA studies, mouse embryonic fibroblasts (MEFs) derived from a homozygous (TG2−/−) mouse contain higher levels of cytochrome c mRNA than those derived from a WT mouse (TG2+/+ MEF).
* Denotes a p < 0.05 and ** denotes a p < 0.01; # or ## denotes a p < 0.05 or <0.01 when Q111 is compared to Q7.

Schematic representation of distinct motifs within the human TG2 sequence heterologously expressed in Q7 and Q111 cells. In order to exclude TG2 from the nucleus, a nuclear export sequence (NES) was introduced at the N-terminal (NES TG2). Additional point mutations were generated in NLS1 (NN TG2). In red, three important amino acids of the transamidating domain; in blue, calcium binding sites; in green, guanine–nucleotide binding site.
Total lysates from Q7 and Q111 cells transfected with WT TG2, or associated NES and NLS mutants. The level of expression is similar in all the samples. β-Actin is used as a loading control. The gel is representative of at least three replicates.
Immunofluorescence staining shows that WT TG2 is expressed in the cytoplasm and nucleus. NES TG2 is primarily expressed in the cytosol, as predicted. Biochemical studies verified the reduced nuclear localization of TG2 with NES addition alone or NES addition plus the NLS 1 mutation (see Supporting Information Fig S6).
Overexpression of NES TG2 and NN TG2 increases the mRNA levels of PGC-1α and cytochrome c genes, compared to WT TG2 in mhtt expressing cells. The results are normalized to WT TG2 expressed in Q7 (n = 6); *p < 0.05 and **p < 0.01.
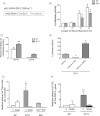
A. A cytochrome c promoter–reporter construct that contains the proximal 326 bp of the cytochrome c promoter (-326-luc), including response elements for specificity protein 1 (SP1), cAMP (CRE) and nuclear respiratory factor 1 (NRF-1).
B. Q7 and Q111 cells transfected with the -326-luc construct followed by various durations of serum starvation and restimulation (serum re-addition). A more robust response of cytochrome c promoter activity is induced within Q7 cells (white bars) than Q111 cells (grey bars) relative to control condition (C) of no serum starvation. These findings indicate that that homeostatic response to energetic stress is repressed in cells expressing mhtt.
C. Co-transfection of a WT TG2 construct with the cytochrome c promoter–reporter greatly inhibited promoter activity in Q7 and Q111 cells compared to cells that were transfected with GFP and the cytochrome c promoter–reporter. Similarly, a TG2 construct that does not retain cross-linking activity (C277S) lost the ability to repress the cytochrome c promoter activity in Q7 or Q111 cells. WT and mutant TG2 were expressed at the same protein levels in these experiments.
D. Inhibition of TG activity by ZDON (50 µM for 12 h) increased the cytochrome c -326 promoter–reporter activity in Q111 cells nearly fourfold over non-treated cells (-326-luc w ZDON). Over-expression of TG2 repressed the promoter activity (-326-luc w TG2).
E, F. Chromatin immunoprecipitation assays showed that more TG2 is located at the cytochrome c promoter (E) and at the PGC-1α gene (F) in non-treated Q111 cells compared to non-treated Q7 cells. TG2 occupancy was sharply decreased in Q111 cells treated with ZDON in both cases. TG2 was found at the cytochrome c promoter of WT MEFs (TG2+/+ MEF) but not in MEFs derived from TG2−/− cells (TG2−/− MEF), confirming the specificity of the TG2 antibody used for ChIP assays. The data representing each sample is normalized with respect to both IgG immunoprecipitations and input.
*p < 0.05 and **p < 0.01.
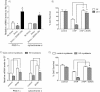
Induction of mhtt by the addition of doxycycline decreased the amount of PGC-1α mRNA in human neuroblastoma cells. This decrease was reversed by 50 µM ZDON. These results demonstrate that silencing of PGC-1α expression occurs very early following mhtt expression, and is not a distal secondary or tertiary effect of chronic mhtt expression. Similarly, TG2 inhibition can nullify these proximate effects of mhtt on gene expression.
Q7 and Q111 cells were protected from 3-NP toxicity (10 mM for 48 h) by pre-treatment with ZDON (50 µM for 12 h). 3-NP inhibits SDH (a TCA cycle and mitochondrial respiratory chain enzyme) to induce death. Note that in agreement with prior studies, Q111 cells are more vulnerable to 3-NP than Q7 cells.
ZDON (50 µM for 2 h) increased the relative amount of PGC-1α and cytochrome c mRNA in both WT human myoblasts and myoblasts derived from HD patients. In accordance with our model, the ZDON-induced protection is highly correlated with reversal of transcriptional dysregulation in HD.
ZDON (50 µM for 12 h) protected myoblasts from WT and HD patients against 3-NP toxicity. These findings suggest that ZDON prevents vulnerability induced by mhtt in a human context.
*p < 0.05.

CS specific activity was increased after 24 h of ZDON treatment. CS is the enzyme that converts pyruvate to Acetyl-CoA is an accepted marker of mitochondrial mass (Chavez et al, 2010).
ND6 was increased relative to β-actin in Q111 cells after 24 h treatment of ZDON. Nuclear encoded mitochondrial transcription factor A (Tfam) is a PGC-1α regulated gene. Tfam, in turn, acts in the mitochondria to induce ND6 expression. The induction of ND6 by ZDON is consistent with ZDON-induced increases in PGC-1α expression and activity.
Mitotracker® staining was increased in both Q7 and Q111 cells after ZDON treatment. Mitochondrial mass is increased by ZDON in both Q7 and Q111 cells. According to the bioenergetic studies the increase in membrane potential could reflect an increase in the biosynthetic capacity of respiratory chain substrates by mitochondrial (TCA cycle) or extramitochondrial sites (pyruvate, fatty acids) by ZDON.
Quantification of Mitotracker® staining with and without the addition of ZDON (50 µM for 12 h).
*p < 0.05 and **p < 0.01.

Q111 cells have 461 dysregulated probes, relative to Q7 cells, and ZDON had a large effect (more than 40% of dysregulated genes) in normalizing this dysfunction. Ontology gene sets affected by ZDON are broad and include p53-regulated genes, glutathione metabolism genes and chaperone genes.
Prototypical HDAC inhibitors NaBu or TSA increased the amount of PGC-1α mRNA in Q111 cells. These findings demonstrate that ZDON induces PCG-1α mRNA expression as well as NaBu or TSA.
ZDON does not affect the acetylation state of H4 as measured by immunoblotting. As expected, the classes I and II HDAC inhibitor TSA increased H4 acetylation. ZDON works independent of canonical changes in histone acetylation to modify transcription. These results are congruent with an HDAC independent role for TG2 in epigenetic regulation.
*p < 0.05 and **p < 0.01.
WT TG2 interacts with Histone H3 in Q7 and Q111 cells supporting the evidence that TG2 modulates transcription. The ability of TG2 to interact with Histone H3 is consistent with a role for TG2 in modulating facultative heterochromatin formation by affecting N-terminal post-transcriptional modifications of histones, also known as the ‘histone code’.

A Drosophila HD model expressing exon 1 of htt with 93 CAG repeats (Q93) fed ZDON (125 µM final concentration in food) displayed a greater number of rhabdomeres per ommatidium than HD Q93 flies receiving no treatment (control) or DMSO alone (Vehicle).
This was calculated as a 21% rescue from DMSO and a 17% rescue from the control group by the formula: 100 × (Rt − Rc)/(7 − Rc), Rt is the number of rhabdomeres/ommatidium in the ZDON group and Rc is the control group.
A Drosophila model of HD fed ZDON (125 µM final concentration in food) showed increased cytochrome c mRNA when compared to those that were not fed ZDON (control) or those fed only DMSO.
*p < 0.05 and **p < 0.01.

TUNEL assay (a biomarker of apoptotic death) of medium-sized spiny neurons following 24 h of NMDA exposure. The MSNs pre-incubated with ZDON (50 µM) for 12 h showed less apoptotic nuclei that the ones untreated or pre-treated with Zc (50 µM), demonstrating the protective effects of TG2 inhibition against NMDA-mediated toxicity. Extrasynaptic NMDA induces changes in cytosolic calcium sufficient to further activate TG2 and repress adaptive transcription. We propose a model whereby TG2 inhibition prevents extrasynaptic glutamate from inducing transcriptional dysregulation and leads to neuroprotection.
Quantification of Fig 8A.
**p < 0.01, comparing WT to Y128; ###p < 0.001, comparing to NMDA treatment group of its genotype.
Comment in
-
Déjà vu with a twist: transglutaminases in bioenergetics and transcriptional dysfunction in Huntington's disease.EMBO Mol Med. 2010 Sep;2(9):335-7. doi: 10.1002/emmm.201000092. EMBO Mol Med. 2010. PMID: 20730854 Free PMC article.
Similar articles
-
Déjà vu with a twist: transglutaminases in bioenergetics and transcriptional dysfunction in Huntington's disease.EMBO Mol Med. 2010 Sep;2(9):335-7. doi: 10.1002/emmm.201000092. EMBO Mol Med. 2010. PMID: 20730854 Free PMC article.
-
Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration.Cell Metab. 2006 Nov;4(5):349-62. doi: 10.1016/j.cmet.2006.10.004. Epub 2006 Oct 19. Cell Metab. 2006. PMID: 17055784
-
Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration.Cell. 2006 Oct 6;127(1):59-69. doi: 10.1016/j.cell.2006.09.015. Cell. 2006. PMID: 17018277
-
PGC-1α at the intersection of bioenergetics regulation and neuron function: from Huntington's disease to Parkinson's disease and beyond.Prog Neurobiol. 2012 May;97(2):142-51. doi: 10.1016/j.pneurobio.2011.10.004. Epub 2011 Nov 9. Prog Neurobiol. 2012. PMID: 22100502 Free PMC article. Review.
-
The interrelationship between mitochondrial dysfunction and transcriptional dysregulation in Huntington disease.J Bioenerg Biomembr. 2010 Jun;42(3):199-205. doi: 10.1007/s10863-010-9286-7. J Bioenerg Biomembr. 2010. PMID: 20556492 Free PMC article. Review.
Cited by
-
Physiological, pathological, and structural implications of non-enzymatic protein-protein interactions of the multifunctional human transglutaminase 2.Cell Mol Life Sci. 2015 Aug;72(16):3009-35. doi: 10.1007/s00018-015-1909-z. Epub 2015 May 6. Cell Mol Life Sci. 2015. PMID: 25943306 Free PMC article. Review.
-
Transglutaminase 2 has opposing roles in the regulation of cellular functions as well as cell growth and death.Cell Death Dis. 2016 Jun 2;7(6):e2244. doi: 10.1038/cddis.2016.150. Cell Death Dis. 2016. PMID: 27253408 Free PMC article. Review.
-
The role of TG2 in regulating S100A4-mediated mammary tumour cell migration.PLoS One. 2013;8(3):e57017. doi: 10.1371/journal.pone.0057017. Epub 2013 Mar 1. PLoS One. 2013. PMID: 23469180 Free PMC article.
-
PGC-1α, mitochondrial dysfunction, and Huntington's disease.Free Radic Biol Med. 2013 Sep;62:37-46. doi: 10.1016/j.freeradbiomed.2013.04.016. Epub 2013 Apr 19. Free Radic Biol Med. 2013. PMID: 23602910 Free PMC article. Review.
-
Transglutaminase 6: a protein associated with central nervous system development and motor function.Amino Acids. 2013 Jan;44(1):161-77. doi: 10.1007/s00726-011-1091-z. Epub 2011 Oct 8. Amino Acids. 2013. PMID: 21984379 Free PMC article.
References
-
- Antonyak MA, Boehm JE, Cerione RA. Phosphoinositide 3-kinase activity is required for retinoic acid-induced expression and activation of the tissue transglutaminase. J Biol Chem. 2002;277:14712–14716. - PubMed
-
- Bailey CD, Johnson GV. The protective effects of cystamine in the R6/2 Huntington's disease mouse involve mechanisms other than the inhibition of tissue transglutaminase. Neurobiol Aging. 2006;27:871–879. - PubMed
-
- Ballestar E, Franco L. Use of the transglutaminase reaction to study the dissociation of histone N-terminal tails from DNA in nucleosome core particles. Biochemistry. 1997;36:5963–5969. - PubMed
-
- Ballestar E, Abad C, Franco L. Core histones are glutaminyl substrates for tissue transglutaminase. J Biol Chem. 1996;271:18817–18824. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

