Genetic control of DH reading frame and its effect on B-cell development and antigen-specifc antibody production
- PMID: 20666706
- PMCID: PMC3676173
- DOI: 10.1615/critrevimmunol.v30.i4.20
Genetic control of DH reading frame and its effect on B-cell development and antigen-specifc antibody production
Abstract
The power of the adaptive immune system to identify novel antigens depends on the ability of lymphocytes to create antigen receptors with diverse antigen-binding sites. For immunoglobulins, CDR (complementarity-determining region)-H3 lies at the center of the antigen-binding site, where it often plays a key role in antigen binding. It is created de novo by VDJ rearrangement and is thus the focus for rearrangement-dependent diversity. CDR-H3 is biased for the inclusion of tyrosine. In seeking to identify the mechanisms controlling CDR-H3 amino acid content, we observed that the coding sequence of DH gene segments demonstrate conservation of reading frame (RF)-specific sequence motifs, with RF1 enriched for tyrosine and depleted of hydrophobic and charged amino acids. Use of DH RF1 in functional VDJ transcripts is preferred from the earliest stages of B-cell development, "pushing" CDR-H3 to include specific categories of tyrosine-enriched antigen-binding sites. With development and maturation, the composition of the CDR-H3 repertoire appears to be pulled into a more refined specific range. Forcing the use of alternative DH RFs by means of gene targeting alters the expressed repertoire, enriching alternative sequence categories. This change in the repertoire variably affects antibody production and the development of specific B-cell subsets.
Figures


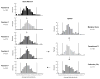


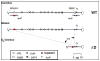


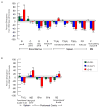

Similar articles
-
HIV-1 gp140 epitope recognition is influenced by immunoglobulin DH gene segment sequence.Immunogenetics. 2016 Feb;68(2):145-55. doi: 10.1007/s00251-015-0890-x. Epub 2015 Dec 19. Immunogenetics. 2016. PMID: 26687685 Free PMC article.
-
The sequences encoded by immunoglobulin diversity (DH ) gene segments play key roles in controlling B-cell development, antigen-binding site diversity, and antibody production.Immunol Rev. 2018 Jul;284(1):106-119. doi: 10.1111/imr.12669. Immunol Rev. 2018. PMID: 29944758 Review.
-
Recirculating bone marrow B cells in C57BL/6 mice are more tolerant of highly hydrophobic and highly charged CDR-H3s than those in BALB/c mice.Eur J Immunol. 2013 Mar;43(3):629-40. doi: 10.1002/eji.201242936. Epub 2013 Jan 18. Eur J Immunol. 2013. PMID: 23225217 Free PMC article.
-
Violation of an evolutionarily conserved immunoglobulin diversity gene sequence preference promotes production of dsDNA-specific IgG antibodies.PLoS One. 2015 Feb 23;10(2):e0118171. doi: 10.1371/journal.pone.0118171. eCollection 2015. PLoS One. 2015. PMID: 25706374 Free PMC article.
-
The role of evolutionarily conserved germ-line DH sequence in B-1 cell development and natural antibody production.Ann N Y Acad Sci. 2015 Dec;1362(1):48-56. doi: 10.1111/nyas.12808. Epub 2015 Jun 23. Ann N Y Acad Sci. 2015. PMID: 26104486 Free PMC article. Review.
Cited by
-
Preimmune Control of the Variance of TCR CDR-B3: Insights Gained From Germline Replacement of a TCR Dβ Gene Segment With an Ig D H Gene Segment.Front Immunol. 2020 Sep 11;11:2079. doi: 10.3389/fimmu.2020.02079. eCollection 2020. Front Immunol. 2020. PMID: 33042119 Free PMC article.
-
Immunoglobulin analysis tool: a novel tool for the analysis of human and mouse heavy and light chain transcripts.Front Immunol. 2012 Jun 28;3:176. doi: 10.3389/fimmu.2012.00176. eCollection 2012. Front Immunol. 2012. PMID: 22754554 Free PMC article.
-
VpreB serves as an invariant surrogate antigen for selecting immunoglobulin antigen-binding sites.Sci Immunol. 2016 Jul 14;1(1):aaf6628. doi: 10.1126/sciimmunol.aaf6628. Sci Immunol. 2016. PMID: 28217764 Free PMC article.
-
Immunoglobulin gene analysis as a tool for investigating human immune responses.Immunol Rev. 2018 Jul;284(1):132-147. doi: 10.1111/imr.12659. Immunol Rev. 2018. PMID: 29944755 Free PMC article. Review.
-
Clonal Progression during the T Cell-Dependent B Cell Antibody Response Depends on the Immunoglobulin DH Gene Segment Repertoire.Front Immunol. 2014 Aug 11;5:385. doi: 10.3389/fimmu.2014.00385. eCollection 2014. Front Immunol. 2014. PMID: 25157256 Free PMC article.
References
-
- Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annual Review of Immunology. 2000;20:197–216. - PubMed
-
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. - PubMed
-
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. - PubMed
-
- Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. 5. U.S. Department of Health and Human Services; Bethesda, Maryland: 1991.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

