Sequence-based identification of interface residues by an integrative profile combining hydrophobic and evolutionary information
- PMID: 20667087
- PMCID: PMC2921408
- DOI: 10.1186/1471-2105-11-402
Sequence-based identification of interface residues by an integrative profile combining hydrophobic and evolutionary information
Abstract
Background: Protein-protein interactions play essential roles in protein function determination and drug design. Numerous methods have been proposed to recognize their interaction sites, however, only a small proportion of protein complexes have been successfully resolved due to the high cost. Therefore, it is important to improve the performance for predicting protein interaction sites based on primary sequence alone.
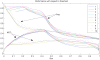
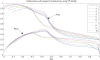
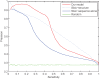
Results: We propose a new idea to construct an integrative profile for each residue in a protein by combining its hydrophobic and evolutionary information. A support vector machine (SVM) ensemble is then developed, where SVMs train on different pairs of positive (interface sites) and negative (non-interface sites) subsets. The subsets having roughly the same sizes are grouped in the order of accessible surface area change before and after complexation. A self-organizing map (SOM) technique is applied to group similar input vectors to make more accurate the identification of interface residues. An ensemble of ten-SVMs achieves an MCC improvement by around 8% and F1 improvement by around 9% over that of three-SVMs. As expected, SVM ensembles constantly perform better than individual SVMs. In addition, the model by the integrative profiles outperforms that based on the sequence profile or the hydropathy scale alone. As our method uses a small number of features to encode the input vectors, our model is simpler, faster and more accurate than the existing methods.
Conclusions: The integrative profile by combining hydrophobic and evolutionary information contributes most to the protein-protein interaction prediction. Results show that evolutionary context of residue with respect to hydrophobicity makes better the identification of protein interface residues. In addition, the ensemble of SVM classifiers improves the prediction performance.
Availability: Datasets and software are available at http://mail.ustc.edu.cn/~bigeagle/BMCBioinfo2010/index.htm.
Figures

Similar articles
-
APIS: accurate prediction of hot spots in protein interfaces by combining protrusion index with solvent accessibility.BMC Bioinformatics. 2010 Apr 8;11:174. doi: 10.1186/1471-2105-11-174. BMC Bioinformatics. 2010. PMID: 20377884 Free PMC article.
-
HemeBIND: a novel method for heme binding residue prediction by combining structural and sequence information.BMC Bioinformatics. 2011 May 26;12:207. doi: 10.1186/1471-2105-12-207. BMC Bioinformatics. 2011. PMID: 21612668 Free PMC article.
-
SVM-Fold: a tool for discriminative multi-class protein fold and superfamily recognition.BMC Bioinformatics. 2007 May 22;8 Suppl 4(Suppl 4):S2. doi: 10.1186/1471-2105-8-S4-S2. BMC Bioinformatics. 2007. PMID: 17570145 Free PMC article.
-
EL_PSSM-RT: DNA-binding residue prediction by integrating ensemble learning with PSSM Relation Transformation.BMC Bioinformatics. 2017 Aug 29;18(1):379. doi: 10.1186/s12859-017-1792-8. BMC Bioinformatics. 2017. PMID: 28851273 Free PMC article.
-
Using residue interaction networks to understand protein function and evolution and to engineer new proteins.Curr Opin Struct Biol. 2024 Dec;89:102922. doi: 10.1016/j.sbi.2024.102922. Epub 2024 Sep 26. Curr Opin Struct Biol. 2024. PMID: 39332048 Free PMC article. Review.
Cited by
-
Prediction of Protein Hotspots from Whole Protein Sequences by a Random Projection Ensemble System.Int J Mol Sci. 2017 Jul 18;18(7):1543. doi: 10.3390/ijms18071543. Int J Mol Sci. 2017. PMID: 28718782 Free PMC article.
-
Prediction of heme binding residues from protein sequences with integrative sequence profiles.Proteome Sci. 2012 Jun 21;10 Suppl 1(Suppl 1):S20. doi: 10.1186/1477-5956-10-S1-S20. Proteome Sci. 2012. PMID: 22759579 Free PMC article.
-
HomPPI: a class of sequence homology based protein-protein interface prediction methods.BMC Bioinformatics. 2011 Jun 17;12:244. doi: 10.1186/1471-2105-12-244. BMC Bioinformatics. 2011. PMID: 21682895 Free PMC article.
-
Unravelling the human taste receptor interactome: machine learning and molecular modelling insights into protein-protein interactions.NPJ Sci Food. 2025 Jul 1;9(1):113. doi: 10.1038/s41538-025-00478-9. NPJ Sci Food. 2025. PMID: 40595706 Free PMC article.
-
DrugECs: An Ensemble System with Feature Subspaces for Accurate Drug-Target Interaction Prediction.Biomed Res Int. 2017;2017:6340316. doi: 10.1155/2017/6340316. Epub 2017 Jul 4. Biomed Res Int. 2017. PMID: 28744468 Free PMC article.
References
-
- Alberts BD, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell. 2. New York: Garland; 1989.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

