Replication-biased genome organisation in the crenarchaeon Sulfolobus
- PMID: 20667100
- PMCID: PMC3091651
- DOI: 10.1186/1471-2164-11-454
Replication-biased genome organisation in the crenarchaeon Sulfolobus
Abstract
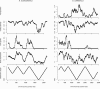
Background: Species of the crenarchaeon Sulfolobus harbour three replication origins in their single circular chromosome that are synchronously initiated during replication.
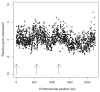
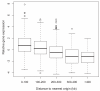
Results: We demonstrate that global gene expression in two Sulfolobus species is highly biased, such that early replicating genome regions are more highly expressed at all three origins. The bias by far exceeds what would be anticipated by gene dosage effects alone. In addition, early replicating regions are denser in archaeal core genes (enriched in essential functions), display lower intergenic distances, and are devoid of mobile genetic elements.
Conclusion: The strong replication-biased structuring of the Sulfolobus chromosome implies that the multiple replication origins serve purposes other than simply shortening the time required for replication. The higher-level chromosomal organisation could be of importance for minimizing the impact of DNA damage, and may also be linked to transcriptional regulation.
Figures
Similar articles
-
Physical and Functional Compartmentalization of Archaeal Chromosomes.Cell. 2019 Sep 19;179(1):165-179.e18. doi: 10.1016/j.cell.2019.08.036. Cell. 2019. PMID: 31539494 Free PMC article.
-
Multiple origins of replication in archaea.Trends Microbiol. 2004 Sep;12(9):399-401. doi: 10.1016/j.tim.2004.07.001. Trends Microbiol. 2004. PMID: 15337158 Review.
-
DNA replication in the hyperthermophilic archaeon Sulfolobus solfataricus.Biochem Soc Trans. 2003 Jun;31(Pt 3):674-6. doi: 10.1042/bst0310674. Biochem Soc Trans. 2003. PMID: 12773180 Review.
-
Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7.DNA Res. 2001 Aug 31;8(4):123-40. doi: 10.1093/dnares/8.4.123. DNA Res. 2001. PMID: 11572479
-
Evolution of the family of pRN plasmids and their integrase-mediated insertion into the chromosome of the crenarchaeon Sulfolobus solfataricus.J Mol Biol. 2000 Nov 3;303(4):449-54. doi: 10.1006/jmbi.2000.4160. J Mol Biol. 2000. PMID: 11054282
Cited by
-
Recombination shapes genome architecture in an organism from the archaeal domain.Genome Biol Evol. 2014 Jan;6(1):170-8. doi: 10.1093/gbe/evu003. Genome Biol Evol. 2014. PMID: 24391154 Free PMC article.
-
Physical and Functional Compartmentalization of Archaeal Chromosomes.Cell. 2019 Sep 19;179(1):165-179.e18. doi: 10.1016/j.cell.2019.08.036. Cell. 2019. PMID: 31539494 Free PMC article.
-
Coevolution of the Organization and Structure of Prokaryotic Genomes.Cold Spring Harb Perspect Biol. 2016 Jan 4;8(1):a018168. doi: 10.1101/cshperspect.a018168. Cold Spring Harb Perspect Biol. 2016. PMID: 26729648 Free PMC article. Review.
-
Non-Random Inversion Landscapes in Prokaryotic Genomes Are Shaped by Heterogeneous Selection Pressures.Mol Biol Evol. 2017 Aug 1;34(8):1902-1911. doi: 10.1093/molbev/msx127. Mol Biol Evol. 2017. PMID: 28407093 Free PMC article.
-
Rings in the extreme: PCNA interactions and adaptations in the archaea.Archaea. 2012;2012:951010. doi: 10.1155/2012/951010. Epub 2012 Nov 14. Archaea. 2012. PMID: 23209375 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

