Norwalk virus does not replicate in human macrophages or dendritic cells derived from the peripheral blood of susceptible humans
- PMID: 20667573
- PMCID: PMC2933743
- DOI: 10.1016/j.virol.2010.07.001
Norwalk virus does not replicate in human macrophages or dendritic cells derived from the peripheral blood of susceptible humans
Abstract
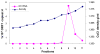
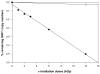
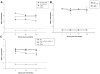
Human noroviruses are difficult to study due to the lack of an efficient in vitro cell culture system or small animal model. Murine norovirus replicates in murine macrophages (MPhi) and dendritic cells (DCs), raising the possibility that human NoVs might replicate in such human cell types. To test this hypothesis, we evaluated DCs and MPhi derived from monocyte subsets and CD11c(+) DCs isolated from peripheral blood mononuclear cells of individuals susceptible to Norwalk virus (NV) infection. These cells were exposed to NV and replication was evaluated by immunofluorescence and by quantitative RT-PCR. A few PBMC-derived DCs expressed NV proteins. However, NV RNA did not increase in any of the cells tested. These results demonstrate that NV does not replicate in human CD11c(+) DCs, monocyte-derived DCs and MPhi, but abortive infection may occur in a few DCs. These results suggest that NV tropism is distinct from that of murine noroviruses.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Norwalk virus RNA is infectious in mammalian cells.J Virol. 2007 Nov;81(22):12238-48. doi: 10.1128/JVI.01489-07. Epub 2007 Sep 12. J Virol. 2007. PMID: 17855551 Free PMC article.
-
Replication and packaging of Norwalk virus RNA in cultured mammalian cells.Proc Natl Acad Sci U S A. 2005 Jul 19;102(29):10327-32. doi: 10.1073/pnas.0408529102. Epub 2005 Jul 7. Proc Natl Acad Sci U S A. 2005. PMID: 16002473 Free PMC article.
-
Evaluation of nine sets of PCR primers in the RNA dependent RNA polymerase region for detection and differentiation of members of the family Caliciviridae, Norwalk virus and Sapporo virus.Microbiol Immunol. 2000;44(5):411-9. doi: 10.1111/j.1348-0421.2000.tb02515.x. Microbiol Immunol. 2000. PMID: 10888362
-
[Norwalk virus: recent findings].Nihon Rinsho. 2002 Jun;60(6):1138-42. Nihon Rinsho. 2002. PMID: 12078086 Review. Japanese.
-
[Rapid and efficient detection method of Norwalk virus].Nihon Rinsho. 2002 Jun;60(6):1181-7. Nihon Rinsho. 2002. PMID: 12078092 Review. Japanese.
Cited by
-
Bile Goes Viral.Viruses. 2021 May 27;13(6):998. doi: 10.3390/v13060998. Viruses. 2021. PMID: 34071855 Free PMC article. Review.
-
Bovine natural antibody IgM inhibits the binding of human norovirus protruding domain to its HBGA receptors.FEBS Open Bio. 2022 Aug;12(8):1489-1497. doi: 10.1002/2211-5463.13450. Epub 2022 Jun 16. FEBS Open Bio. 2022. PMID: 35674188 Free PMC article.
-
In Vitro Replication of Human Norovirus.Viruses. 2019 Jun 12;11(6):547. doi: 10.3390/v11060547. Viruses. 2019. PMID: 31212759 Free PMC article. Review.
-
Differential regulation of CD103 (αE integrin) expression in human dendritic cells by retinoic acid and Toll-like receptor ligands.J Leukoc Biol. 2017 May;101(5):1169-1180. doi: 10.1189/jlb.1MA0316-131R. Epub 2017 Jan 13. J Leukoc Biol. 2017. PMID: 28087652 Free PMC article.
-
B cell lines fail to support efficient rhesus enteric calicivirus and human norovirus replication.J Virol. 2025 May 20;99(5):e0014325. doi: 10.1128/jvi.00143-25. Epub 2025 Apr 22. J Virol. 2025. PMID: 40261012 Free PMC article.
References
-
- Blutt SE, Conner ME. Rotavirus: to the gut and beyond! Curr Opin Gastroenterol. 2007;23:39–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

