NMR structure and ion channel activity of the p7 protein from hepatitis C virus
- PMID: 20667830
- PMCID: PMC2951219
- DOI: 10.1074/jbc.M110.122895
NMR structure and ion channel activity of the p7 protein from hepatitis C virus
Abstract
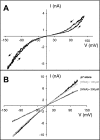
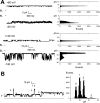
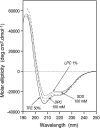
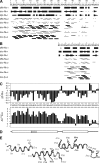
The small membrane protein p7 of hepatitis C virus forms oligomers and exhibits ion channel activity essential for virus infectivity. These viroporin features render p7 an attractive target for antiviral drug development. In this study, p7 from strain HCV-J (genotype 1b) was chemically synthesized and purified for ion channel activity measurements and structure analyses. p7 forms cation-selective ion channels in planar lipid bilayers and at the single-channel level by the patch clamp technique. Ion channel activity was shown to be inhibited by hexamethylene amiloride but not by amantadine. Circular dichroism analyses revealed that the structure of p7 is mainly α-helical, irrespective of the membrane mimetic medium (e.g. lysolipids, detergents, or organic solvent/water mixtures). The secondary structure elements of the monomeric form of p7 were determined by (1)H and (13)C NMR in trifluoroethanol/water mixtures. Molecular dynamics simulations in a model membrane were combined synergistically with structural data obtained from NMR experiments. This approach allowed us to determine the secondary structure elements of p7, which significantly differ from predictions, and to propose a three-dimensional model of the monomeric form of p7 associated with the phospholipid bilayer. These studies revealed the presence of a turn connecting an unexpected N-terminal α-helix to the first transmembrane helix, TM1, and a long cytosolic loop bearing the dibasic motif and connecting TM1 to TM2. These results provide the first detailed experimental structural framework for a better understanding of p7 processing, oligomerization, and ion channel gating mechanism.
Figures

References
-
- Poynard T., Yuen M. F., Ratziu V., Lai C. L. (2003) Lancet 362, 2095–2100 - PubMed
-
- Simmonds P., Bukh J., Combet C., Deléage G., Enomoto N., Feinstone S., Halfon P., Inchauspé G., Kuiken C., Maertens G., Mizokami M., Murphy D. G., Okamoto H., Pawlotsky J. M., Penin F., Sablon E., Shin-I T., Stuyver L. J., Thiel H. J., Viazov S., Weiner A. J., Widell A. (2005) Hepatology 42, 962–973 - PubMed
-
- Lindenbach B. D., Thiel H. J., Rice C. M. (2007) in Fields Virology, 5th Ed., Vol. 1 (Knipe D. M., Howley P. M. eds) pp. 1101–1152, Lippincott-Raven Publishers, Philadelphia, PA
-
- Moradpour D., Penin F., Rice C. M. (2007) Nat. Rev. Microbiol. 5, 453–463 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

