Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
- PMID: 20667835
- PMCID: PMC2945551
- DOI: 10.1074/jbc.M110.111856
Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
Abstract
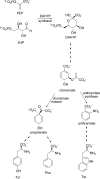
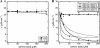
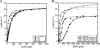
The shikimate pathway, responsible for aromatic amino acid biosynthesis, is required for the growth of Mycobacterium tuberculosis and is a potential drug target. The first reaction is catalyzed by 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (DAH7PS). Feedback regulation of DAH7PS activity by aromatic amino acids controls shikimate pathway flux. Whereas Mycobacterium tuberculosis DAH7PS (MtuDAH7PS) is not inhibited by the addition of Phe, Tyr, or Trp alone, combinations cause significant loss of enzyme activity. In the presence of 200 μm Phe, only 2.4 μm Trp is required to reduce enzymic activity to 50%. Reaction kinetics were analyzed in the presence of inhibitory concentrations of Trp/Phe or Trp/Tyr. In the absence of inhibitors, the enzyme follows Michaelis-Menten kinetics with respect to substrate erythrose 4-phosphate (E4P), whereas the addition of inhibitor combinations caused significant homotropic cooperativity with respect to E4P, with Hill coefficients of 3.3 (Trp/Phe) and 2.8 (Trp/Tyr). Structures of MtuDAH7PS/Trp/Phe, MtuDAH7PS/Trp, and MtuDAH7PS/Phe complexes were determined. The MtuDAH7PS/Trp/Phe homotetramer binds four Trp and six Phe molecules. Binding sites for both aromatic amino acids are formed by accessory elements to the core DAH7PS (β/α)(8) barrel that are unique to the type II DAH7PS family and contribute to the tight dimer and tetramer interfaces. A comparison of the liganded and unliganded MtuDAH7PS structures reveals changes in the interface areas associated with inhibitor binding and a small displacement of the E4P binding loop. These studies uncover a previously unrecognized mode of control for the branched pathways of aromatic amino acid biosynthesis involving synergistic inhibition by specific pairs of pathway end products.
Figures
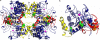

References
-
- Bentley R. (1990) Crit. Rev. Biochem. Mol. Biol. 25, 307–384 - PubMed
-
- Walsh C. T., Liu J., Rusnak F., Sakaitani M. (1990) Chem. Rev. 90, 1105–1129
-
- Roberts F., Roberts C. W., Johnson J. J., Kyle D. E., Krell T., Coggins J. R., Coombs G. H., Milhous W. K., Tzipori S., Ferguson D. J., Chakrabarti D., McLeod R. (1998) Nature 393, 801–805 - PubMed
-
- Campbell S. A., Richards T. A., Mui E. J., Samuel B. U., Coggins J. R., McLeod R., Roberts C. W. (2004) Int. J. Parasitol. 34, 5–13 - PubMed
-
- Parish T., Stoker N. G. (2002) Microbiology 148, 3069–3077 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

