P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages
- PMID: 20668078
- PMCID: PMC2937795
- DOI: 10.1128/JVI.00585-10
P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages
Abstract
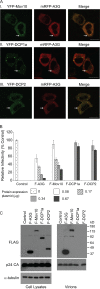
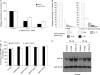
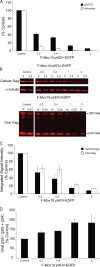
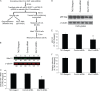
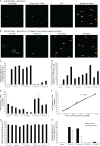
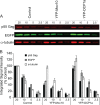
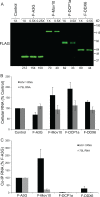
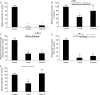
Recent studies have shown that APOBEC3G (A3G), a potent inhibitor of human immunodeficiency virus type 1 (HIV-1) replication, is localized to cytoplasmic mRNA-processing bodies (P bodies). However, the functional relevance of A3G colocalization with P body marker proteins has not been established. To explore the relationship between HIV-1, A3G, and P bodies, we analyzed the effects of overexpression of P body marker proteins Mov10, DCP1a, and DCP2 on HIV-1 replication. Our results show that overexpression of Mov10, a putative RNA helicase that was previously reported to belong to the DExD superfamily and was recently reported to belong to the Upf1-like group of helicases, but not the decapping enzymes DCP1a and DCP2, leads to potent inhibition of HIV-1 replication at multiple stages. Mov10 overexpression in the virus producer cells resulted in reductions in the steady-state levels of the HIV-1 Gag protein and virus production; Mov10 was efficiently incorporated into virions and reduced virus infectivity, in part by inhibiting reverse transcription. In addition, A3G and Mov10 overexpression reduced proteolytic processing of HIV-1 Gag. The inhibitory effects of A3G and Mov10 were additive, implying a lack of functional interaction between the two inhibitors. Small interfering RNA (siRNA)-mediated knockdown of endogenous Mov10 by 80% resulted in a 2-fold reduction in virus production but no discernible impact on the infectivity of the viruses after normalization for the p24 input, suggesting that endogenous Mov10 was not required for viral infectivity. Overall, these results show that Mov10 can potently inhibit HIV-1 replication at multiple stages.
Figures
Similar articles
-
Mov10 and APOBEC3G localization to processing bodies is not required for virion incorporation and antiviral activity.J Virol. 2013 Oct;87(20):11047-62. doi: 10.1128/JVI.02070-13. Epub 2013 Aug 7. J Virol. 2013. PMID: 23926332 Free PMC article.
-
Moloney leukemia virus 10 (MOV10) inhibits the degradation of APOBEC3G through interference with the Vif-mediated ubiquitin-proteasome pathway.Retrovirology. 2017 Dec 19;14(1):56. doi: 10.1186/s12977-017-0382-1. Retrovirology. 2017. PMID: 29258557 Free PMC article.
-
Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses.Retrovirology. 2012 Jun 22;9:53. doi: 10.1186/1742-4690-9-53. Retrovirology. 2012. PMID: 22727223 Free PMC article.
-
Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC-3G and APOBEC-3F restriction factors.Microbiol Mol Biol Rev. 2009 Jun;73(2):211-32. doi: 10.1128/MMBR.00040-08. Microbiol Mol Biol Rev. 2009. PMID: 19487726 Free PMC article. Review.
-
Apobec3G-Based Strategies to Defeat HIV Infection.Curr HIV Res. 2016;14(3):217-24. doi: 10.2174/1570162x14999160224100541. Curr HIV Res. 2016. PMID: 26957196 Review.
Cited by
-
Sequence requirements for localization and packaging of Ty3 retroelement RNA.Virus Res. 2013 Feb;171(2):319-31. doi: 10.1016/j.virusres.2012.10.008. Epub 2012 Oct 13. Virus Res. 2013. PMID: 23073180 Free PMC article.
-
APOBEC3 inhibits DEAD-END function to regulate microRNA activity.BMC Mol Biol. 2013 Jul 26;14:16. doi: 10.1186/1471-2199-14-16. BMC Mol Biol. 2013. PMID: 23890083 Free PMC article.
-
HELZ directly interacts with CCR4-NOT and causes decay of bound mRNAs.Life Sci Alliance. 2019 Sep 30;2(5):e201900405. doi: 10.26508/lsa.201900405. Print 2019 Oct. Life Sci Alliance. 2019. PMID: 31570513 Free PMC article.
-
Comparative Description of the Expression Profile of Interferon-Stimulated Genes in Multiple Cell Lineages Targeted by HIV-1 Infection.Front Microbiol. 2019 Mar 12;10:429. doi: 10.3389/fmicb.2019.00429. eCollection 2019. Front Microbiol. 2019. PMID: 30915053 Free PMC article.
-
Unwinding the roles of RNA helicase MOV10.Wiley Interdiscip Rev RNA. 2022 Mar;13(2):e1682. doi: 10.1002/wrna.1682. Epub 2021 Jul 29. Wiley Interdiscip Rev RNA. 2022. PMID: 34327836 Free PMC article. Review.
References
-
- Brass, A. L., D. M. Dykxhoorn, Y. Benita, N. Yan, A. Engelman, R. J. Xavier, J. Lieberman, and S. J. Elledge. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921-926. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

