Proteolytic products of the porcine reproductive and respiratory syndrome virus nsp2 replicase protein
- PMID: 20668084
- PMCID: PMC2937792
- DOI: 10.1128/JVI.01208-10
Proteolytic products of the porcine reproductive and respiratory syndrome virus nsp2 replicase protein
Abstract
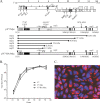
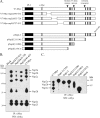
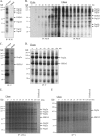
The nsp2 replicase protein of porcine reproductive and respiratory syndrome virus (PRRSV) was recently demonstrated to be processed from its precursor by the PL2 protease at or near the G(1196)|G(1197) dipeptide in transfected CHO cells. Here the proteolytic cleavage of PRRSV nsp2 was further investigated in virally infected MARC-145 cells by using two recombinant PRRSVs expressing epitope-tagged nsp2. The data revealed that PRRSV nsp2 exists as different isoforms, termed nsp2a, nsp2b, nsp2c, nsp2d, nsp2e, and nsp2f, during PRRSV infection. Moreover, on the basis of deletion mutagenesis and antibody probing, these nsp2 species appeared to share the same N terminus but to differ in their C termini. The largest protein, nsp2a, corresponded to the nsp2 product identified in transfected CHO cells. nsp2b and nsp2c were processed within or near the transmembrane (TM) region, presumably at or near the conserved sites G(981)|G(982) and G(828)|G(829)|G(830), respectively. The C termini for nsp2d, -e, and -f were mapped within the nsp2 middle hypervariable region, but no conserved cleavage sites could be definitively predicted. The larger nsp2 species emerged almost simultaneously in the early stage of PRRSV infection. Pulse-chase analysis revealed that all six nsp2 species were relatively stable and had low turnover rates. Deletion mutagenesis revealed that the smaller nsp2 species (e.g., nsp2d, nsp2e, and nsp2f) were not essential for viral replication in cell culture. Lastly, we identified a cellular chaperone, named heat shock 70-kDa protein 5 (HSPA5), that was strongly associated with nsp2, which may have important implications for PRRSV replication. Overall, these findings indicate that PRRSV nsp2 is increasingly emerging as a multifunctional protein and may have a profound impact on PRRSV replication and viral pathogenesis.
Figures
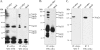


Similar articles
-
The nsp2 Hypervariable Region of Porcine Reproductive and Respiratory Syndrome Virus Strain JXwn06 Is Associated with Viral Cellular Tropism to Primary Porcine Alveolar Macrophages.J Virol. 2019 Nov 26;93(24):e01436-19. doi: 10.1128/JVI.01436-19. Print 2019 Dec 15. J Virol. 2019. PMID: 31554681 Free PMC article.
-
Identification of nonessential regions of the nsp2 replicase protein of porcine reproductive and respiratory syndrome virus strain VR-2332 for replication in cell culture.J Virol. 2007 Sep;81(18):9878-90. doi: 10.1128/JVI.00562-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522233 Free PMC article.
-
Proteolytic processing of the porcine reproductive and respiratory syndrome virus replicase.Virus Res. 2015 Apr 16;202:48-59. doi: 10.1016/j.virusres.2014.12.027. Epub 2014 Dec 31. Virus Res. 2015. PMID: 25557977
-
The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins.Virus Res. 2010 Dec;154(1-2):61-76. doi: 10.1016/j.virusres.2010.07.030. Epub 2010 Aug 7. Virus Res. 2010. PMID: 20696193 Free PMC article. Review.
-
In vivo growth of porcine reproductive and respiratory syndrome virus engineered nsp2 deletion mutants.Virus Res. 2010 Dec;154(1-2):77-85. doi: 10.1016/j.virusres.2010.07.024. Epub 2010 Jul 29. Virus Res. 2010. PMID: 20673840 Free PMC article. Review.
Cited by
-
Construction of a Porcine Reproductive and Respiratory Syndrome Virus with Nanoluc Luciferase Reporter: a Stable and Highly Efficient Tool for Viral Quantification Both In Vitro and In Vivo.Microbiol Spectr. 2022 Aug 31;10(4):e0027622. doi: 10.1128/spectrum.00276-22. Epub 2022 Jun 27. Microbiol Spectr. 2022. PMID: 35758677 Free PMC article.
-
Interleukin-2 enhancer binding factor 2 interacts with the nsp9 or nsp2 of porcine reproductive and respiratory syndrome virus and exerts negatively regulatory effect on the viral replication.Virol J. 2017 Jul 11;14(1):125. doi: 10.1186/s12985-017-0794-5. Virol J. 2017. PMID: 28693575 Free PMC article.
-
Porcine Reproductive and Respiratory Syndrome Virus: Immune Escape and Application of Reverse Genetics in Attenuated Live Vaccine Development.Vaccines (Basel). 2021 May 10;9(5):480. doi: 10.3390/vaccines9050480. Vaccines (Basel). 2021. PMID: 34068505 Free PMC article. Review.
-
Porcine reproductive and respiratory syndrome virus nonstructural protein 2 contributes to NF-κB activation.Virol J. 2012 Apr 30;9:83. doi: 10.1186/1743-422X-9-83. Virol J. 2012. PMID: 22546080 Free PMC article.
-
Molecular characterization of a complete genome and 12 Nsp2 genes of PRRSV of southwestern China.Food Environ Virol. 2012 Sep;4(3):102-14. doi: 10.1007/s12560-012-9083-z. Epub 2012 Jul 12. Food Environ Virol. 2012. PMID: 23412837
References
-
- Castón, J. R., J. L. Martinez-Torrecuadrada, A. Maraver, E. Lombardo, J. F. Rodriguez, J. I. Casal, and J. L. Carrascosa. 2001. C terminus of infectious bursal disease virus major capsid protein VP2 is involved in definition of the T number for capsid assembly. J. Virol. 75:10815-10828. - PMC - PubMed
-
- Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

