A new cistron in the murine hepatitis virus replicase gene
- PMID: 20668085
- PMCID: PMC2937787
- DOI: 10.1128/JVI.00901-10
A new cistron in the murine hepatitis virus replicase gene
Abstract
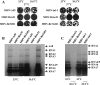
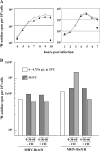
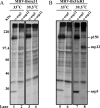
We report an RNA-negative, temperature-sensitive (ts) mutant of Murine hepatitis virus, Bristol ts31 (MHV-Brts31), that defines a new complementation group within the MHV replicase gene locus. MHV-Brts31 has near-normal levels of RNA synthesis at the permissive temperature of 33 degrees C but is unable to synthesize viral RNA when the infection is initiated and maintained at the nonpermissive temperature of 39.5 degrees C. Sequence analysis of MHV-Brts31 RNA indicated that a single G-to-A transition at codon 1307 in open reading frame 1a, which results in a replacement of methionine-475 with isoleucine in nonstructural protein 3 (nsp3), was responsible for the ts phenotype. This conclusion was confirmed using a vaccinia virus-based reverse genetics system to produce a recombinant virus, Bristol tsc31 (MHV-Brtsc31), which has the same RNA-negative ts phenotype and complementation profile as those of MHV-Brts31. The analysis of protein synthesis in virus-infected cells showed that, at the nonpermissive temperature, MHV-Brtsc31 was not able to proteolytically process either p150, the precursor polypeptide of the replicase nonstructural proteins nsp4 to nsp10, or the replicase polyprotein pp1ab to produce nsp12. The processing of replicase polyprotein pp1a in the region of nsp1 to nsp3 was not affected. Transmission electron microscopy showed that, compared to revertant virus, the number of double-membrane vesicles in MHV-Brts31-infected cells is reduced at the nonpermissive temperature. These results identify a new cistron in the MHV replicase gene locus and show that nsp3 has an essential role in the assembly of a functional MHV replication-transcription complex.
Figures
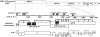


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

