Lipids and proteins act in opposing manners to regulate polyomavirus infection
- PMID: 20668088
- PMCID: PMC2937789
- DOI: 10.1128/JVI.01093-10
Lipids and proteins act in opposing manners to regulate polyomavirus infection
Abstract
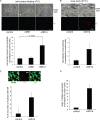
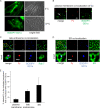
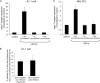
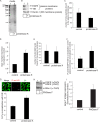
How receptors control virus infection is poorly understood. Polyomavirus (Py) binds to the sialic acid-galactose moiety on receptors to gain entry into host cells and cause infection. We previously demonstrated that the sialic acid-galactose-containing glycolipids called gangliosides GD1a and GT1b promote Py infection, in part, by sorting the virus from the endolysosomes to the endoplasmic reticulum (ER), a critical infection route. Whether these glycolipids act as Py entry receptors, however, is not clear. Additionally, as the majority of glycoproteins also harbor terminal sialic acid-galactose residues, their roles in Py infection are also not well established. Using a ganglioside-deficient cell line, we show that GD1a is the functional entry receptor for Py. GD1a binds to Py on the plasma membrane, and the receptor-virus complex is internalized and transported to the late endosomes and then the ER to initiate infection. In contrast, our findings indicate that glycoproteins act as decoy receptors, restricting the ER transport and infection of Py. Thus, glycolipids and glycoproteins, two major constituents of the plasma membrane, execute opposing functions in regulating infection by a defined virus.
Figures

Similar articles
-
A lipid receptor sorts polyomavirus from the endolysosome to the endoplasmic reticulum to cause infection.PLoS Pathog. 2009 Jun;5(6):e1000465. doi: 10.1371/journal.ppat.1000465. Epub 2009 Jun 5. PLoS Pathog. 2009. PMID: 19503604 Free PMC article.
-
Gangliosides are receptors for murine polyoma virus and SV40.EMBO J. 2003 Sep 1;22(17):4346-55. doi: 10.1093/emboj/cdg439. EMBO J. 2003. PMID: 12941687 Free PMC article.
-
Ganglioside GD1a restores infectibility to mouse cells lacking functional receptors for polyomavirus.J Virol. 2005 Jan;79(1):615-8. doi: 10.1128/JVI.79.1.615-618.2005. J Virol. 2005. PMID: 15596855 Free PMC article.
-
The Polyomaviridae: Contributions of virus structure to our understanding of virus receptors and infectious entry.Virology. 2009 Feb 20;384(2):389-99. doi: 10.1016/j.virol.2008.12.021. Epub 2009 Jan 21. Virology. 2009. PMID: 19157478 Free PMC article. Review.
-
SV40 Hijacks Cellular Transport, Membrane Penetration, and Disassembly Machineries to Promote Infection.Viruses. 2019 Oct 5;11(10):917. doi: 10.3390/v11100917. Viruses. 2019. PMID: 31590347 Free PMC article. Review.
Cited by
-
Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain.Clin Microbiol Rev. 2012 Jul;25(3):471-506. doi: 10.1128/CMR.05031-11. Clin Microbiol Rev. 2012. PMID: 22763635 Free PMC article. Review.
-
Virus recognition of glycan receptors.Curr Opin Virol. 2019 Feb;34:117-129. doi: 10.1016/j.coviro.2019.01.004. Epub 2019 Mar 5. Curr Opin Virol. 2019. PMID: 30849709 Free PMC article. Review.
-
JC Polyomavirus Attachment and Entry: Potential Sites for PML Therapeutics.Curr Clin Microbiol Rep. 2017 Sep;4(3):132-141. doi: 10.1007/s40588-017-0069-3. Epub 2017 Aug 1. Curr Clin Microbiol Rep. 2017. PMID: 28989857 Free PMC article.
-
Virus-Receptor Interactions: The Key to Cellular Invasion.J Mol Biol. 2018 Aug 17;430(17):2590-2611. doi: 10.1016/j.jmb.2018.06.024. Epub 2018 Jun 18. J Mol Biol. 2018. PMID: 29924965 Free PMC article. Review.
-
α2-3- and α2-6- N-linked sialic acids allow efficient interaction of Newcastle Disease Virus with target cells.Glycoconj J. 2012 Oct;29(7):539-49. doi: 10.1007/s10719-012-9431-0. Epub 2012 Aug 7. Glycoconj J. 2012. PMID: 22869099 Free PMC article.
References
-
- Baranowski, E., C. M. Ruiz-Jarabo, and E. Domingo. 2001. Evolution of cell recognition by viruses. Science 292:1102-1105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

