Cellular microRNAs 200b and 429 regulate the Epstein-Barr virus switch between latency and lytic replication
- PMID: 20668090
- PMCID: PMC2937814
- DOI: 10.1128/JVI.00923-10
Cellular microRNAs 200b and 429 regulate the Epstein-Barr virus switch between latency and lytic replication
Abstract
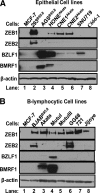
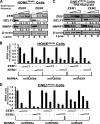
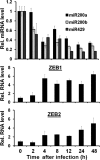
We previously showed that the cellular proteins ZEB1 and ZEB2/SIP1 both play key roles in regulating the latent-lytic switch of Epstein-Barr Virus (EBV) by repressing BZLF1 gene expression. We investigated here the effects of cellular microRNA (miRNA) 200 (miR200) family members on the EBV infection status of cells. We show that miR200b and miR429, but not miR200a, can induce EBV-positive cells into lytic replication by downregulating expression of ZEB1 and ZEB2, leading to production of infectious virus. The levels of miR200 family members in EBV-infected cells strongly negatively correlated with the levels of the ZEBs (e.g., -0.89 [P < 0.001] for miR429 versus ZEB1) and positively correlated with the degree of EBV lytic gene expression (e.g., 0.73 [P < 0.01] for miR429 versus BZLF1). The addition of either miR200b or miR429 to EBV-positive cells led to EBV lytic reactivation in a ZEB-dependent manner; inhibition of these miRNAs led to decreased EBV lytic gene expression. The degree of latent infection by an EBV mutant defective in the primary ZEB-binding site of the EBV BZLF1 promoter was not affected by the addition of these miRNAs. Furthermore, EBV infection of primary blood B cells led to downregulation of these miRNAs and upregulation of ZEB levels. Thus, we conclude that miRNAs 200b and 429 are key regulators via their effects on expression of ZEB1 and ZEB2 of the switch between latent and lytic infection by EBV and, therefore, potential targets for development of new lytic induction therapeutics with which to treat patients with EBV-associated malignancies.
Figures










Similar articles
-
Either ZEB1 or ZEB2/SIP1 can play a central role in regulating the Epstein-Barr virus latent-lytic switch in a cell-type-specific manner.J Virol. 2010 Jun;84(12):6139-52. doi: 10.1128/JVI.02706-09. Epub 2010 Apr 7. J Virol. 2010. PMID: 20375168 Free PMC article.
-
ZEB1 regulates the latent-lytic switch in infection by Epstein-Barr virus.PLoS Pathog. 2007 Dec;3(12):e194. doi: 10.1371/journal.ppat.0030194. PLoS Pathog. 2007. PMID: 18085824 Free PMC article.
-
Zinc finger E-box binding factor 1 plays a central role in regulating Epstein-Barr virus (EBV) latent-lytic switch and acts as a therapeutic target in EBV-associated gastric cancer.Cancer. 2012 Feb 15;118(4):924-36. doi: 10.1002/cncr.26184. Epub 2011 Jun 29. Cancer. 2012. PMID: 21717425
-
Latent and lytic Epstein-Barr virus replication strategies.Rev Med Virol. 2005 Jan-Feb;15(1):3-15. doi: 10.1002/rmv.441. Rev Med Virol. 2005. PMID: 15386591 Review.
-
Switching of EBV cycles between latent and lytic states.Rev Med Virol. 2014 May;24(3):142-53. doi: 10.1002/rmv.1780. Epub 2013 Dec 11. Rev Med Virol. 2014. PMID: 24339346 Review.
Cited by
-
B Cell Receptor-Responsive miR-141 Enhances Epstein-Barr Virus Lytic Cycle via FOXO3 Inhibition.mSphere. 2021 Apr 14;6(2):e00093-21. doi: 10.1128/mSphere.00093-21. mSphere. 2021. PMID: 33853871 Free PMC article.
-
miRNAs: The Key Regulator of COVID-19 Disease.Int J Cell Biol. 2022 Oct 29;2022:1645366. doi: 10.1155/2022/1645366. eCollection 2022. Int J Cell Biol. 2022. PMID: 36345541 Free PMC article. Review.
-
Involvement of eukaryotic small RNA pathways in host defense and viral pathogenesis.Viruses. 2013 Oct 30;5(11):2659-78. doi: 10.3390/v5112659. Viruses. 2013. PMID: 24178713 Free PMC article. Review.
-
MiR-10a-5p-Mediated Syndecan 1 Suppression Restricts Porcine Hemagglutinating Encephalomyelitis Virus Replication.Front Microbiol. 2020 Feb 20;11:105. doi: 10.3389/fmicb.2020.00105. eCollection 2020. Front Microbiol. 2020. PMID: 32153518 Free PMC article.
-
Epstein-Barr Virus-associated lymphoproliferative disorders: experimental and clinical developments.Int J Clin Exp Med. 2015 Sep 15;8(9):14656-71. eCollection 2015. Int J Clin Exp Med. 2015. PMID: 26628948 Free PMC article. Review.
References
-
- Adam, L., M. Zhong, W. Choi, W. Qi, M. Nicoloso, A. Arora, G. Calin, H. Wang, A. Siefker-Radtke, D. McConkey, M. Bar-Eli, and C. Dinney. 2009. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin. Cancer Res. 15:5060-5072. - PMC - PubMed
-
- Ambros, V. 2000. Control of developmental timing in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 10:428-433. - PubMed
-
- Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

