Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments
- PMID: 20668487
- PMCID: PMC3105697
- DOI: 10.1038/ismej.2010.117
Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments
Abstract
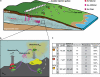
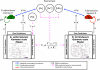
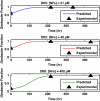
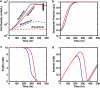
The advent of rapid complete genome sequencing, and the potential to capture this information in genome-scale metabolic models, provide the possibility of comprehensively modeling microbial community interactions. For example, Rhodoferax and Geobacter species are acetate-oxidizing Fe(III)-reducers that compete in anoxic subsurface environments and this competition may have an influence on the in situ bioremediation of uranium-contaminated groundwater. Therefore, genome-scale models of Geobacter sulfurreducens and Rhodoferax ferrireducens were used to evaluate how Geobacter and Rhodoferax species might compete under diverse conditions found in a uranium-contaminated aquifer in Rifle, CO. The model predicted that at the low rates of acetate flux expected under natural conditions at the site, Rhodoferax will outcompete Geobacter as long as sufficient ammonium is available. The model also predicted that when high concentrations of acetate are added during in situ bioremediation, Geobacter species would predominate, consistent with field-scale observations. This can be attributed to the higher expected growth yields of Rhodoferax and the ability of Geobacter to fix nitrogen. The modeling predicted relative proportions of Geobacter and Rhodoferax in geochemically distinct zones of the Rifle site that were comparable to those that were previously documented with molecular techniques. The model also predicted that under nitrogen fixation, higher carbon and electron fluxes would be diverted toward respiration rather than biomass formation in Geobacter, providing a potential explanation for enhanced in situ U(VI) reduction in low-ammonium zones. These results show that genome-scale modeling can be a useful tool for predicting microbial interactions in subsurface environments and shows promise for designing bioremediation strategies.
Figures



References
-
- Anesiadis N, Cluett WR, Mahadevan R. Dynamic metabolic engineering for increasing bioprocess productivity. Metab Eng. 2008;10:255–266. - PubMed
-
- Balba MT, Nedwell DB. Microbial metabolism of acetate, propionate and butyrate in anoxic sediment from the Colne Point Saltmarsh, Essex, UK. J Gen Microbiol. 1982;128:1415–1422.
-
- Burgard AP, Pharkya P, Maranas CD. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84:647–657. - PubMed

