Rab32 modulates apoptosis onset and mitochondria-associated membrane (MAM) properties
- PMID: 20670942
- PMCID: PMC2951233
- DOI: 10.1074/jbc.M110.101584
Rab32 modulates apoptosis onset and mitochondria-associated membrane (MAM) properties
Abstract
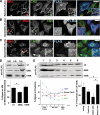
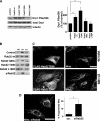
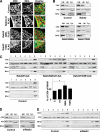
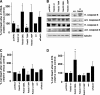
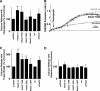
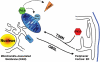
The mitochondria-associated membrane (MAM) has emerged as an endoplasmic reticulum (ER) signaling hub that accommodates ER chaperones, including the lectin calnexin. At the MAM, these chaperones control ER homeostasis but also play a role in the onset of ER stress-mediated apoptosis, likely through the modulation of ER calcium signaling. These opposing roles of MAM-localized chaperones suggest the existence of mechanisms that regulate the composition and the properties of ER membrane domains. Our results now show that the GTPase Rab32 localizes to the ER and mitochondria, and we identify this protein as a regulator of MAM properties. Consistent with such a role, Rab32 modulates ER calcium handling and disrupts the specific enrichment of calnexin on the MAM, while not affecting the ER distribution of protein-disulfide isomerase and mitofusin-2. Furthermore, Rab32 determines the targeting of PKA to mitochondrial and ER membranes and through its overexpression or inactivation increases the phosphorylation of Bad and of Drp1. Through a combination of its functions as a PKA-anchoring protein and a regulator of MAM properties, the activity and expression level of Rab32 determine the speed of apoptosis onset.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

