Proper levels of the Arabidopsis cohesion establishment factor CTF7 are essential for embryo and megagametophyte, but not endosperm, development
- PMID: 20671110
- PMCID: PMC2949036
- DOI: 10.1104/pp.110.157560
Proper levels of the Arabidopsis cohesion establishment factor CTF7 are essential for embryo and megagametophyte, but not endosperm, development
Abstract
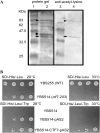
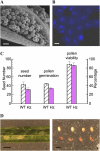
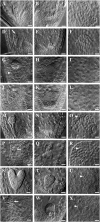
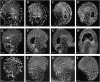
CTF7 is an essential gene in yeast that is required for the formation of sister chromatid cohesion. While recent studies have provided insights into how sister chromatid cohesion is established, less is known about how specifically CTF7 facilitates the formation of cohesion, and essentially nothing is known about how sister chromatid cohesion is established in plants. In this report, we describe the isolation and characterization of CTF7 from Arabidopsis (Arabidopsis thaliana). Arabidopsis CTF7 is similar to Saccharomyces cerevisiae CTF7 in that it lacks an amino-terminal extension, exhibits acetyltransferase activity, and can complement a yeast ctf7 temperature-sensitive mutation. CTF7 transcripts are found throughout the plant, with the highest levels present in buds. Seeds containing T-DNA insertions in CTF7 exhibit mitotic defects in the zygote. Interestingly, the endosperm developed normally in ctf7 seeds, suggesting that CTF7 is not essential for mitosis in endosperm nuclei. Minor defects were observed in female gametophytes of ctf7(+/-) plants, and plants that overexpress CTF7 exhibited female gametophyte lethality. Pollen development appeared normal in both CTF7 knockout and overexpression plants. Therefore, proper levels of CTF7 are critical for female gametophyte and embryo development but not for the establishment of mitotic cohesion during microgametogenesis or during endosperm development.
Figures


Similar articles
-
AtCTF7 is required for establishment of sister chromatid cohesion and association of cohesin with chromatin during meiosis in Arabidopsis.BMC Plant Biol. 2013 Aug 14;13:117. doi: 10.1186/1471-2229-13-117. BMC Plant Biol. 2013. PMID: 23941555 Free PMC article.
-
Arabidopsis CHROMOSOME TRANSMISSION FIDELITY 7 (AtCTF7/ECO1) is required for DNA repair, mitosis and meiosis.Plant J. 2013 Sep;75(6):927-40. doi: 10.1111/tpj.12261. Epub 2013 Jul 20. Plant J. 2013. PMID: 23750584 Free PMC article.
-
The Opposing Actions of Arabidopsis CHROMOSOME TRANSMISSION FIDELITY7 and WINGS APART-LIKE1 and 2 Differ in Mitotic and Meiotic Cells.Plant Cell. 2016 Feb;28(2):521-36. doi: 10.1105/tpc.15.00781. Epub 2016 Jan 26. Plant Cell. 2016. PMID: 26813623 Free PMC article.
-
Cloning of Xenopus orthologs of Ctf7/Eco1 acetyltransferase and initial characterization of XEco2.FEBS J. 2008 Dec;275(24):6109-22. doi: 10.1111/j.1742-4658.2008.06736.x. Epub 2008 Oct 30. FEBS J. 2008. PMID: 19016859
-
The long and winding road: transport pathways for amino acids in Arabidopsis seeds.Plant Reprod. 2018 Sep;31(3):253-261. doi: 10.1007/s00497-018-0334-5. Epub 2018 Mar 16. Plant Reprod. 2018. PMID: 29549431 Review.
Cited by
-
AtCTF7 is required for establishment of sister chromatid cohesion and association of cohesin with chromatin during meiosis in Arabidopsis.BMC Plant Biol. 2013 Aug 14;13:117. doi: 10.1186/1471-2229-13-117. BMC Plant Biol. 2013. PMID: 23941555 Free PMC article.
-
Arabidopsis CHROMOSOME TRANSMISSION FIDELITY 7 (AtCTF7/ECO1) is required for DNA repair, mitosis and meiosis.Plant J. 2013 Sep;75(6):927-40. doi: 10.1111/tpj.12261. Epub 2013 Jul 20. Plant J. 2013. PMID: 23750584 Free PMC article.
-
Involvement of the Cohesin Cofactor PDS5 (SPO76) During Meiosis and DNA Repair in Arabidopsis thaliana.Front Plant Sci. 2015 Dec 1;6:1034. doi: 10.3389/fpls.2015.01034. eCollection 2015. Front Plant Sci. 2015. PMID: 26648949 Free PMC article.
-
Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA.Plant Mol Biol. 2014 Aug;85(6):541-50. doi: 10.1007/s11103-014-0202-0. Epub 2014 May 30. Plant Mol Biol. 2014. PMID: 24874772 Free PMC article.
-
Overexpression of a truncated CTF7 construct leads to pleiotropic defects in reproduction and vegetative growth in Arabidopsis.BMC Plant Biol. 2015 Mar 5;15:74. doi: 10.1186/s12870-015-0452-2. BMC Plant Biol. 2015. PMID: 25848842 Free PMC article.
References
-
- An Y, McDowell J, Huang S, McKinney E, Chambliss S, Meagher R. (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 - PubMed
-
- Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. (2008) Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321: 563–566 - PubMed
-
- Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C. (1999) The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 19: 463–472 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

