Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells
- PMID: 20671182
- PMCID: PMC2922915
- DOI: 10.1126/science.1188308
Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells
Erratum in
- Science. 2011 Oct 28;334(6055):453
Abstract
Protein and messenger RNA (mRNA) copy numbers vary from cell to cell in isogenic bacterial populations. However, these molecules often exist in low copy numbers and are difficult to detect in single cells. We carried out quantitative system-wide analyses of protein and mRNA expression in individual cells with single-molecule sensitivity using a newly constructed yellow fluorescent protein fusion library for Escherichia coli. We found that almost all protein number distributions can be described by the gamma distribution with two fitting parameters which, at low expression levels, have clear physical interpretations as the transcription rate and protein burst size. At high expression levels, the distributions are dominated by extrinsic noise. We found that a single cell's protein and mRNA copy numbers for any given gene are uncorrelated.
Figures

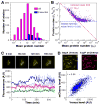

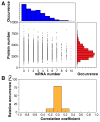

Comment in
-
Genomics. E. coli, what a noisy bug.Science. 2010 Jul 30;329(5991):518-9. doi: 10.1126/science.1194036. Science. 2010. PMID: 20671174 No abstract available.
-
Single molecule: Single molecules meet systems biology.Nat Methods. 2010 Sep;7(9):672-3. doi: 10.1038/nmeth0910-672a. Nat Methods. 2010. PMID: 20827801
References
-
- Rao CV, Wolf DM, Arkin AP. Nature. 2002 Nov 14;420:231. - PubMed
-
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Science. 2005 Mar 25;307:1962. - PubMed
-
- Pedraza JM, van Oudenaarden A. Science. 2005 Mar 25;307:1965. - PubMed
-
- Friedman N, Cai L, Xie XS. Physical review letters. 2006 Oct 20;97:168302. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

