Nucleotide supply, not local histone acetylation, sets replication origin usage in transcribed regions
- PMID: 20671737
- PMCID: PMC2933864
- DOI: 10.1038/embor.2010.112
Nucleotide supply, not local histone acetylation, sets replication origin usage in transcribed regions
Abstract
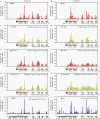
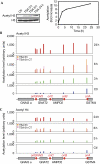
In eukaryotes, only a fraction of replication origins fire at each S phase. Local histone acetylation was proposed to control firing efficiency of origins, but conflicting results were obtained. We report that local histone acetylation does not reflect origin efficiencies along the adenosine monophosphate deaminase 2 locus in mammalian fibroblasts. Reciprocally, modulation of origin efficiency does not affect acetylation. However, treatment with a deacetylase inhibitor changes the initiation pattern. We demonstrate that this treatment alters pyrimidine biosynthesis and decreases fork speed, which recruits latent origins. Our findings reconcile results that seemed inconsistent and reveal an unsuspected effect of deacetylase inhibitors on replication dynamics.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Aggarwal BD, Calvi BR (2004) Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376 - PubMed
-
- Anglana M, Apiou F, Bensimon A, Debatisse M (2003) Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 - PubMed
-
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M (2008) Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455: 557–560 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

