Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development
- PMID: 20671958
- PMCID: PMC2910479
- DOI: 10.1155/2010/297505
Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development
Abstract
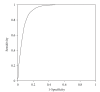
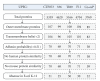
Vaxign is the first web-based vaccine design system that predicts vaccine targets based on genome sequences using the strategy of reverse vaccinology. Predicted features in the Vaxign pipeline include protein subcellular location, transmembrane helices, adhesin probability, conservation to human and/or mouse proteins, sequence exclusion from genome(s) of nonpathogenic strain(s), and epitope binding to MHC class I and class II. The precomputed Vaxign database contains prediction of vaccine targets for >70 genomes. Vaxign also performs dynamic vaccine target prediction based on input sequences. To demonstrate the utility of this program, the vaccine candidates against uropathogenic Escherichia coli (UPEC) were predicted using Vaxign and compared with various experimental studies. Our results indicate that Vaxign is an accurate and efficient vaccine design program.
Figures


References
-
- Rappuoli R. Reverse vaccinology. Current Opinion in Microbiology. 2000;3(5):445–450. - PubMed
-
- Pizza M, Scarlato V, Masignani V, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816–1820. - PubMed
-
- Ross BC, Czajkowski L, Hocking D, et al. Identification of vaccine candidate antigens from a genomic analysis of Porphyromonas gingivalis. Vaccine. 2001;19(30):4135–4142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

