A Comprehensive Study of Progressive Cytogenetic Alterations in Clear Cell Renal Cell Carcinoma and a New Model for ccRCC Tumorigenesis and Progression
- PMID: 20671976
- PMCID: PMC2909727
- DOI: 10.1155/2010/428325
A Comprehensive Study of Progressive Cytogenetic Alterations in Clear Cell Renal Cell Carcinoma and a New Model for ccRCC Tumorigenesis and Progression
Abstract
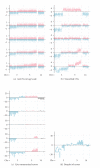
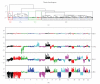
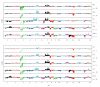
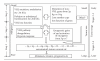
We present a comprehensive study of cytogenetic alterations that occur during the progression of clear cell renal cell carcinoma (ccRCC). We used high-density high-throughput Affymetrix 100 K SNP arrays to obtain the whole genome SNP copy number information from 71 pretreatment tissue samples with RCC tumors; of those, 42 samples were of human ccRCC subtype. We analyzed patterns of cytogenetic loss and gain from different RCC subtypes and in particular, different stages and grades of ccRCC tumors, using a novel algorithm that we have designed. Based on patterns of cytogenetic alterations in chromosomal regions with frequent losses and gains, we inferred the involvement of candidate genes from these regions in ccRCC tumorigenesis and development. We then proposed a new model of ccRCC tumorigenesis and progression. Our study serves as a comprehensive overview of cytogenetic alterations in a collection of 572 ccRCC tumors from diversified studies and should facilitate the search for specific genes associated with the disease.
Figures

References
-
- Tan M-H, Rogers CG, Cooper JT, et al. Gene expression profiling of renal cell carcinoma. Clinical Cancer Research. 2004;10(18, part 2):6315S–6321S. - PubMed
-
- Cohen HT, McGovern FJ. Renal-cell carcinoma. The New England Journal of Medicine. 2005;353(23):2477–2490. - PubMed
-
- Latif F, Tory K, Gnarra J, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. The Science. 1993;260(5112):1317–1320. - PubMed
-
- Skinnider BF, Folpe AL, Hennigar RA, et al. Distribution of cytokeratins and vimentin in adult renal neoplasms and normal renal tissue: potential utility of a cytokeratin antibody panel in the differential diagnosis of renal tumors. American Journal of Surgical Pathology. 2005;29(6):747–754. - PubMed
LinkOut - more resources
Full Text Sources

