Conformational dynamics of actin: effectors and implications for biological function
- PMID: 20672362
- PMCID: PMC3038201
- DOI: 10.1002/cm.20473
Conformational dynamics of actin: effectors and implications for biological function
Abstract
Actin is a protein abundant in many cell types. Decades of investigations have provided evidence that it has many functions in living cells. The diverse morphology and dynamics of actin structures adapted to versatile cellular functions is established by a large repertoire of actin-binding proteins. The proper interactions with these proteins assume effective molecular adaptations from actin, in which its conformational transitions play essential role. This review attempts to summarise our current knowledge regarding the coupling between the conformational states of actin and its biological function.
Figures


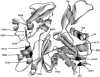
Similar articles
-
The actin cytoskeleton of Dictyostelium: a story told by mutants.J Cell Sci. 2000 Mar;113 ( Pt 5):759-66. doi: 10.1242/jcs.113.5.759. J Cell Sci. 2000. PMID: 10671366 Review.
-
Regulation of the cytoskeleton assembly: a role for a ternary complex of actin with two actin-binding proteins.Results Probl Cell Differ. 2001;32:165-79. doi: 10.1007/978-3-540-46560-7_12. Results Probl Cell Differ. 2001. PMID: 11131830 Review. No abstract available.
-
Regulation of actin cytoskeleton dynamics in cells.Mol Cells. 2010 Apr;29(4):311-25. doi: 10.1007/s10059-010-0053-8. Mol Cells. 2010. PMID: 20446344 Free PMC article. Review.
-
Signaling to the actin cytoskeleton.Annu Rev Cell Dev Biol. 1998;14:305-38. doi: 10.1146/annurev.cellbio.14.1.305. Annu Rev Cell Dev Biol. 1998. PMID: 9891786 Review.
-
Cytoskeleton: actin and endocytosis--no longer the weakest link.Curr Biol. 2001 Sep 4;11(17):R691-4. doi: 10.1016/s0960-9822(01)00410-9. Curr Biol. 2001. PMID: 11553341 Review.
Cited by
-
MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation.Mol Cell. 2013 Aug 8;51(3):397-404. doi: 10.1016/j.molcel.2013.06.019. Epub 2013 Aug 1. Mol Cell. 2013. PMID: 23911929 Free PMC article.
-
Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection.Biochim Biophys Acta Bioenerg. 2017 Aug;1858(8):602-614. doi: 10.1016/j.bbabio.2017.01.004. Epub 2017 Jan 16. Biochim Biophys Acta Bioenerg. 2017. PMID: 28104365 Free PMC article. Review.
-
Myosin and tropomyosin stabilize the conformation of formin-nucleated actin filaments.J Biol Chem. 2012 Sep 14;287(38):31894-904. doi: 10.1074/jbc.M112.341230. Epub 2012 Jun 29. J Biol Chem. 2012. PMID: 22753415 Free PMC article.
-
Actin polymerization negatively regulates p53 function by impairing its nuclear import in response to DNA damage.PLoS One. 2013;8(4):e60179. doi: 10.1371/journal.pone.0060179. Epub 2013 Apr 2. PLoS One. 2013. PMID: 23565200 Free PMC article.
-
Ethanol Effect on BK Channels is Modulated by Magnesium.Alcohol Clin Exp Res. 2015 Sep;39(9):1671-9. doi: 10.1111/acer.12821. Alcohol Clin Exp Res. 2015. PMID: 26331878 Free PMC article.
References
-
- Allen PG, Laham LE, Way M, Janmey PA. Binding of phosphate, aluminum fluoride, or beryllium fluoride to F-actin inhibits severing by gelsolin. J Biol Chem. 1996;271:4665–4670. - PubMed
-
- Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. - PubMed
-
- Arii Y, Hatori K. Relationship between the flexibility and the motility of actin filaments: effects of pH. Biochem Biophys Res Commun. 2008;371:772–776. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

