The Drosophila homolog of the mammalian imprint regulator, CTCF, maintains the maternal genomic imprint in Drosophila melanogaster
- PMID: 20673338
- PMCID: PMC2922095
- DOI: 10.1186/1741-7007-8-105
The Drosophila homolog of the mammalian imprint regulator, CTCF, maintains the maternal genomic imprint in Drosophila melanogaster
Abstract
Background: CTCF is a versatile zinc finger DNA-binding protein that functions as a highly conserved epigenetic transcriptional regulator. CTCF is known to act as a chromosomal insulator, bind promoter regions, and facilitate long-range chromatin interactions. In mammals, CTCF is active in the regulatory regions of some genes that exhibit genomic imprinting, acting as insulator on only one parental allele to facilitate parent-specific expression. In Drosophila, CTCF acts as a chromatin insulator and is thought to be actively involved in the global organization of the genome.
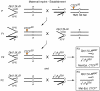
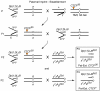
Results: To determine whether CTCF regulates imprinting in Drosophila, we generated CTCF mutant alleles and assayed gene expression from the imprinted Dp(1;f)LJ9 mini-X chromosome in the presence of reduced CTCF expression. We observed disruption of the maternal imprint when CTCF levels were reduced, but no effect was observed on the paternal imprint. The effect was restricted to maintenance of the imprint and was specific for the Dp(1;f)LJ9 mini-X chromosome.
Conclusions: CTCF in Drosophila functions in maintaining parent-specific expression from an imprinted domain as it does in mammals. We propose that Drosophila CTCF maintains an insulator boundary on the maternal X chromosome, shielding genes from the imprint-induced silencing that occurs on the paternally inherited X chromosome. See commentary: http://www.biomedcentral.com/1741-7007/8/104.
Figures






Comment in
-
Insulators and imprinting from flies to mammals.BMC Biol. 2010 Jul 30;8:104. doi: 10.1186/1741-7007-8-104. BMC Biol. 2010. PMID: 20687908 Free PMC article.
Similar articles
-
Genomic imprinting in Drosophila is maintained by the products of Suppressor of variegation and trithorax group, but not Polycomb group, genes.Mol Genet Genomics. 2002 Sep;268(1):103-12. doi: 10.1007/s00438-002-0731-0. Epub 2002 Aug 7. Mol Genet Genomics. 2002. PMID: 12242505
-
Insulators and imprinting from flies to mammals.BMC Biol. 2010 Jul 30;8:104. doi: 10.1186/1741-7007-8-104. BMC Biol. 2010. PMID: 20687908 Free PMC article.
-
The Insulator Protein CTCF Is Required for Correct Hox Gene Expression, but Not for Embryonic Development in Drosophila.Genetics. 2018 Sep;210(1):129-136. doi: 10.1534/genetics.118.301350. Epub 2018 Jul 18. Genetics. 2018. PMID: 30021792 Free PMC article.
-
Imprinting of the mouse Igf2r gene depends on an intronic CpG island.Mol Cell Endocrinol. 1998 May 25;140(1-2):9-14. doi: 10.1016/s0303-7207(98)00022-7. Mol Cell Endocrinol. 1998. PMID: 9722161 Review.
-
Differential 3D chromatin organization and gene activity in genomic imprinting.Curr Opin Genet Dev. 2020 Apr;61:17-24. doi: 10.1016/j.gde.2020.03.004. Epub 2020 Apr 13. Curr Opin Genet Dev. 2020. PMID: 32299027 Review.
Cited by
-
Insights into HP1a-Chromatin Interactions.Cells. 2020 Aug 9;9(8):1866. doi: 10.3390/cells9081866. Cells. 2020. PMID: 32784937 Free PMC article. Review.
-
Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster.BMC Biol. 2015 Aug 7;13:63. doi: 10.1186/s12915-015-0168-7. BMC Biol. 2015. PMID: 26248466 Free PMC article.
-
Epigenetic mechanisms of genomic imprinting: common themes in the regulation of imprinted regions in mammals, plants, and insects.Genet Res Int. 2012;2012:585024. doi: 10.1155/2012/585024. Epub 2012 Feb 15. Genet Res Int. 2012. PMID: 22567394 Free PMC article.
-
Exome sequencing identifies a recurrent de novo ZSWIM6 mutation associated with acromelic frontonasal dysostosis.Am J Hum Genet. 2014 Aug 7;95(2):235-40. doi: 10.1016/j.ajhg.2014.07.008. Am J Hum Genet. 2014. PMID: 25105228 Free PMC article.
-
Genomic imprinting absent in Drosophila melanogaster adult females.Cell Rep. 2012 Jul 26;2(1):69-75. doi: 10.1016/j.celrep.2012.06.013. Epub 2012 Jul 20. Cell Rep. 2012. PMID: 22840398 Free PMC article.
References
-
- Filippova G. Genetics and epigenetics of the multifunctional protein CTCF. Curr Top in Dev Biol. 2008;80:337–360. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Mukhopadhyay R, Yu W, Whitehead J, Xu J, Lezcano M, Pack S, Kanduri C, Kanduri M, Ginjala V, Vostrov A, Quitschke W, Chernukhin I, Klenova E, Lobanenkov V, Ohlsson R. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 2004;14:1594–1602. - PMC - PubMed
-
- Wan L-B, Bartolomei MS. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv Genet. 2008;61:207–223. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

