Predicting beta-turns and their types using predicted backbone dihedral angles and secondary structures
- PMID: 20673368
- PMCID: PMC2920885
- DOI: 10.1186/1471-2105-11-407
Predicting beta-turns and their types using predicted backbone dihedral angles and secondary structures
Abstract
Background: Beta-turns are secondary structure elements usually classified as coil. Their prediction is important, because of their role in protein folding and their frequent occurrence in protein chains.
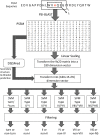
Results: We have developed a novel method that predicts beta-turns and their types using information from multiple sequence alignments, predicted secondary structures and, for the first time, predicted dihedral angles. Our method uses support vector machines, a supervised classification technique, and is trained and tested on three established datasets of 426, 547 and 823 protein chains. We achieve a Matthews correlation coefficient of up to 0.49, when predicting the location of beta-turns, the highest reported value to date. Moreover, the additional dihedral information improves the prediction of beta-turn types I, II, IV, VIII and "non-specific", achieving correlation coefficients up to 0.39, 0.33, 0.27, 0.14 and 0.38, respectively. Our results are more accurate than other methods.
Conclusions: We have created an accurate predictor of beta-turns and their types. Our method, called DEBT, is available online at http://comp.chem.nottingham.ac.uk/debt/.
Figures


Similar articles
-
Prediction of backbone dihedral angles and protein secondary structure using support vector machines.BMC Bioinformatics. 2009 Dec 22;10:437. doi: 10.1186/1471-2105-10-437. BMC Bioinformatics. 2009. PMID: 20025785 Free PMC article.
-
Prediction of beta-turns at over 80% accuracy based on an ensemble of predicted secondary structures and multiple alignments.BMC Bioinformatics. 2008 Oct 10;9:430. doi: 10.1186/1471-2105-9-430. BMC Bioinformatics. 2008. PMID: 18847492 Free PMC article.
-
NetTurnP--neural network prediction of beta-turns by use of evolutionary information and predicted protein sequence features.PLoS One. 2010 Nov 30;5(11):e15079. doi: 10.1371/journal.pone.0015079. PLoS One. 2010. PMID: 21152409 Free PMC article.
-
[A turning point in the knowledge of the structure-function-activity relations of elastin].J Soc Biol. 2001;195(2):181-93. J Soc Biol. 2001. PMID: 11727705 Review. French.
-
Computational methods for protein secondary structure prediction using multiple sequence alignments.Curr Protein Pept Sci. 2000 Nov;1(3):273-301. doi: 10.2174/1389203003381324. Curr Protein Pept Sci. 2000. PMID: 12369910 Review.
Cited by
-
RaptorX-Angle: real-value prediction of protein backbone dihedral angles through a hybrid method of clustering and deep learning.BMC Bioinformatics. 2018 May 8;19(Suppl 4):100. doi: 10.1186/s12859-018-2065-x. BMC Bioinformatics. 2018. PMID: 29745828 Free PMC article.
-
A new clustering and nomenclature for beta turns derived from high-resolution protein structures.PLoS Comput Biol. 2019 Mar 7;15(3):e1006844. doi: 10.1371/journal.pcbi.1006844. eCollection 2019 Mar. PLoS Comput Biol. 2019. PMID: 30845191 Free PMC article.
-
ccPDB 2.0: an updated version of datasets created and compiled from Protein Data Bank.Database (Oxford). 2019 Jan 1;2019:bay142. doi: 10.1093/database/bay142. Database (Oxford). 2019. PMID: 30689843 Free PMC article.
-
A deep dense inception network for protein beta-turn prediction.Proteins. 2020 Jan;88(1):143-151. doi: 10.1002/prot.25780. Epub 2019 Jul 23. Proteins. 2020. PMID: 31294886 Free PMC article.
-
Evaluation of protein dihedral angle prediction methods.PLoS One. 2014 Aug 28;9(8):e105667. doi: 10.1371/journal.pone.0105667. eCollection 2014. PLoS One. 2014. PMID: 25166857 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

