Genomic repertoires of DNA-binding transcription factors across the tree of life
- PMID: 20675356
- PMCID: PMC2995046
- DOI: 10.1093/nar/gkq617
Genomic repertoires of DNA-binding transcription factors across the tree of life
Abstract
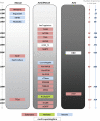
Sequence-specific transcription factors (TFs) are important to genetic regulation in all organisms because they recognize and directly bind to regulatory regions on DNA. Here, we survey and summarize the TF resources available. We outline the organisms for which TF annotation is provided, and discuss the criteria and methods used to annotate TFs by different databases. By using genomic TF repertoires from ∼700 genomes across the tree of life, covering Bacteria, Archaea and Eukaryota, we review TF abundance with respect to the number of genes, as well as their structural complexity in diverse lineages. While typical eukaryotic TFs are longer than the average eukaryotic proteins, the inverse is true for prokaryotes. Only in eukaryotes does the same family of DNA-binding domain (DBD) occur multiple times within one polypeptide chain. This potentially increases the length and diversity of DNA-recognition sequence by reusing DBDs from the same family. We examined the increase in TF abundance with the number of genes in genomes, using the largest set of prokaryotic and eukaryotic genomes to date. As pointed out before, prokaryotic TFs increase faster than linearly. We further observe a similar relationship in eukaryotic genomes with a slower increase in TFs.
Figures

References
-
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. - PubMed
-
- Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. - PubMed
-
- Carroll SB. Endless forms: the evolution of gene regulation and morphological diversity. Cell. 2000;101:577–580. - PubMed
-
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. - PubMed
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

