A genomic portrait of human microsatellite variation
- PMID: 20675409
- PMCID: PMC3002246
- DOI: 10.1093/molbev/msq198
A genomic portrait of human microsatellite variation
Abstract
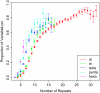
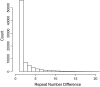
Rapid advances in DNA sequencing and genotyping technologies are beginning to reveal the scope and pattern of human genomic variation. Although single nucleotide polymorphisms (SNPs) have been intensively studied, the extent and form of variation at other types of molecular variants remain poorly understood. Polymorphism at the most variable loci in the human genome, microsatellites, has rarely been examined on a genomic scale without the ascertainment biases that attend typical genotyping studies. We conducted a genomic survey of variation at microsatellites with at least three perfect repeats by comparing two complete genome sequences, the Human Genome Reference sequence and the sequence of J. Craig Venter. The genomic proportion of polymorphic loci was 2.7%, much higher than the rate of SNP variation, with marked heterogeneity among classes of loci. The proportion of variable loci increased substantially with repeat number. Repeat lengths differed in levels of variation, with longer repeat lengths generally showing higher polymorphism at the same repeat number. Microsatellite variation was weakly correlated with regional SNP number, indicating modest effects of shared genealogical history. Reductions in variation were detected at microsatellites located in introns, in untranslated regions, in coding exons, and just upstream of transcription start sites, suggesting the presence of selective constraints. Our results provide new insights into microsatellite mutational processes and yield a preview of patterns of variation that will be obtained in genomic surveys of larger numbers of individuals.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

