Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production
- PMID: 20675495
- PMCID: PMC2944552
- DOI: 10.1128/JB.00445-10
Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production
Abstract
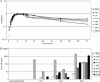
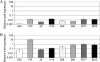
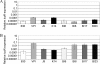
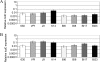
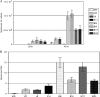
Toxigenic Clostridium difficile strains produce two toxins (TcdA and TcdB) during the stationary phase of growth and are the leading cause of antibiotic-associated diarrhea. C. difficile isolates of the molecular type NAP1/027/BI have been associated with severe disease and hospital outbreaks worldwide. It has been suggested that these "hypervirulent" strains produce larger amounts of toxin and that a mutation in a putative negative regulator (TcdC) allows toxin production at all growth phases. To rigorously explore this possibility, we conducted a quantitative examination of the toxin production of multiple hypervirulent and nonhypervirulent C. difficile strains. Toxin gene (tcdA and tcdB) and toxin gene regulator (tcdR and tcdC) expression was also monitored. To obtain additional correlates for the hypervirulence phenotype, sporulation kinetics and efficiency were measured. In the exponential phase, low basal levels of tcdA, tcdB, and tcdR expression were evident in both hypervirulent and nonhypervirulent strains, but contrary to previous assumptions, toxin levels were below the detectable thresholds. While hypervirulent strains displayed robust toxin production during the stationary phase of growth, the amounts were not significantly different from those of the nonhypervirulent strains tested; further, total toxin amounts were directly proportional to tcdA, tcdB, and tcdR gene expression. Interestingly, tcdC expression did not diminish in stationary phase, suggesting that TcdC may have a modulatory rather than a strictly repressive role. Comparative genomic analyses of the closely related nonhypervirulent strains VPI 10463 (the highest toxin producer) and 630 (the lowest toxin producer) revealed polymorphisms in the tcdR ribosome binding site and the tcdR-tcdB intergenic region, suggesting that a mechanistic basis for increased toxin production in VPI 10463 could be increased TcdR translation and read-through transcription of the tcdA and tcdB genes. Hypervirulent isolates produced significantly more spores, and did so earlier, than all other isolates. Increased sporulation, potentially in synergy with robust toxin production, may therefore contribute to the widespread disease now associated with hypervirulent C. difficile strains.
Figures

References
-
- Akerlund, T., B. Svenungsson, A. Lagergren, and L. G. Burman. 2006. Correlation of disease severity with fecal toxin levels in patients with Clostridium difficile-associated diarrhea and distribution of PCR ribotypes and toxin yields in vitro of corresponding isolates. J. Clin. Microbiol. 44:353-358. - PMC - PubMed
-
- Alfa, M. J., C. Dueck, N. Olson, P. Degagne, S. Papetti, A. Wald, E. Lo, and G. Harding. 2008. UV-visible marker confirms that environmental persistence of Clostridium difficile spores in toilets of patients with C. difficile-associated diarrhea is associated with lack of compliance with cleaning protocol. BMC Infect. Dis. 8:64. - PMC - PubMed
-
- Aziz, R. K., D. Bartels, A. A. Best, M. DeJongh, T. Disz, R. A. Edwards, K. Formsma, S. Gerdes, E. M. Glass, M. Kubal, F. Meyer, G. J. Olsen, R. Olson, A. L. Osterman, R. A. Overbeek, L. K. McNeil, D. Parrmann, T. Paczian, B. Parrello, G. D. Pusch, C. Reich, R. Stevens, O. Vassieva, V. Vonstein, A. Wilke, and I. Zagnitko. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75-89. - PMC - PubMed
-
- Barbut, F., B. Gariazzo, L. Bonne, V. Lalande, B. Burghoffer, R. Luiuz, and J. C. Petit. 2007. Clinical features of Clostridium difficile-associated infections and molecular characterization of strains: results of a retrospective study, 2000-2004. Infect. Control Hosp. Epidemiol. 28:131-139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

