The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12
- PMID: 20675712
- PMCID: PMC2951861
- DOI: 10.1093/hmg/ddq324
The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12
Abstract
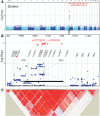
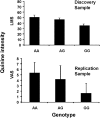
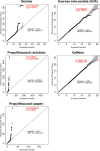
The perceived taste intensities of quinine HCl, caffeine, sucrose octaacetate (SOA) and propylthiouracil (PROP) solutions were examined in 1457 twins and their siblings. Previous heritability modeling of these bitter stimuli indicated a common genetic factor for quinine, caffeine and SOA (22-28%), as well as separate specific genetic factors for PROP (72%) and quinine (15%). To identify the genes involved, we performed a genome-wide association study with the same sample as the modeling analysis, genotyped for approximately 610,000 single-nucleotide polymorphisms (SNPs). For caffeine and SOA, no SNP association reached a genome-wide statistical criterion. For PROP, the peak association was within TAS2R38 (rs713598, A49P, P = 1.6 × 10(-104)), which accounted for 45.9% of the trait variance. For quinine, the peak association was centered in a region that contains bitter receptor as well as salivary protein genes and explained 5.8% of the trait variance (TAS2R19, rs10772420, R299C, P = 1.8 × 10(-15)). We confirmed this association in a replication sample of twins of similar ancestry (P = 0.00001). The specific genetic factor for the perceived intensity of PROP was identified as the gene previously implicated in this trait (TAS2R38). For quinine, one or more bitter receptor or salivary proline-rich protein genes on chromosome 12 have alleles which affect its perception but tight linkage among very similar genes precludes the identification of a single causal genetic variant.
Figures

References
-
- Falconer D.S. Sensory thresholds for solutions of phenyl-thio-carbamide. Ann. Eugen. 1947;13:211–222. - PubMed
-
- Fischer R., Griffin F. Quinine dimorphism: a cardinal determinant of taste sensitivity. Nature. 1963;200:343–347. doi:10.1038/200343a0. - DOI - PubMed
-
- Smith S.E., Davies P.D. Quinine taste thresholds: a family study and a twin study. Ann. Hum. Genet. 1973;37:227–232. doi:10.1111/j.1469-1809.1973.tb01830.x. - DOI - PubMed
-
- Hansen J.L., Reed D.R., Wright M.J., Martin N.G., Breslin P.A. Heritability and genetic covariation of sensitivity to PROP, SOA, quinine HCl, and caffeine. Chem. Senses. 2006;31:403–413. doi:10.1093/chemse/bjj044. - DOI - PMC - PubMed
-
- Kim U.K., Jorgenson E., Coon H., Leppert M., Risch N., Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi:10.1126/science.1080190. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

