Dynamics of a novel centromeric histone variant CenH3 reveals the evolutionary ancestral timing of centromere biogenesis
- PMID: 20675719
- PMCID: PMC2995078
- DOI: 10.1093/nar/gkq664
Dynamics of a novel centromeric histone variant CenH3 reveals the evolutionary ancestral timing of centromere biogenesis
Abstract
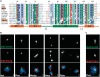
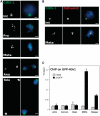
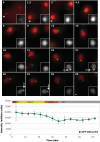
The centromeric histone H3 variant (CenH3) serves to target the kinetochore to the centromeres and thus ensures correct chromosome segregation during mitosis and meiosis. The Dictyostelium H3-like variant H3v1 was identified as the CenH3 ortholog. Dictyostelium CenH3 has an extended N-terminal domain with no similarity to any other known proteins and a histone fold domain at its C-terminus. Within the histone fold, α-helix 2 (α2) and an extended loop 1 (L1) have been shown to be required for targeting CenH3 to centromeres. Compared to other known and putative CenH3 histones, Dictyostelium CenH3 has a shorter L1, suggesting that the extension is not an obligatory feature. Through ChIP analysis and fluorescence microscopy of live and fixed cells, we provide here the first survey of centromere structure in amoebozoa. The six telocentric centromeres were found to mostly consist of all the DIRS-1 elements and to associate with H3K9me3. During interphase, the centromeres remain attached to the centrosome forming a single CenH3-containing cluster. Loading of Dictyostelium CenH3 onto centromeres occurs at the G2/prophase transition, in contrast to the anaphase/telophase loading of CenH3 observed in metazoans. This suggests that loading during G2/prophase is the ancestral eukaryotic mechanism and that anaphase/telophase loading of CenH3 has evolved more recently after the amoebozoa diverged from the animal linage.
Figures


References
-
- Gieni RS, Chan GK, Hendzel MJ. Epigenetics regulate centromere formation and kinetochore function. J. Cellular Biochem. 2008;104:2027–2039. - PubMed
-
- Tyler-Smith C, Floridia G. Many paths to the top of the mountain, diverse evolutionary solutions to centromere structure. Cell. 2000;102:5–8. - PubMed
-
- Ekwall K. Epigenetic control of centromere behavior. Annu. Rev. Genet. 2007;41:63–81. - PubMed
-
- Cooper JL, Henikoff S. Adaptive evolution of the histone fold domain in centromeric histones. Mol. Biol. Evol. 2004;21:1712–1718. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

