HEXIM1 targets a repeated GAUC motif in the riboregulator of transcription 7SK and promotes base pair rearrangements
- PMID: 20675720
- PMCID: PMC2995076
- DOI: 10.1093/nar/gkq660
HEXIM1 targets a repeated GAUC motif in the riboregulator of transcription 7SK and promotes base pair rearrangements
Abstract
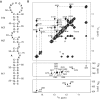
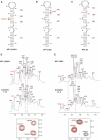
7SK snRNA, an abundant RNA discovered in human nucleus, regulates transcription by RNA polymerase II (RNAPII). It sequesters and inhibits the transcription elongation factor P-TEFb which, by phosphorylation of RNAPII, switches transcription from initiation to processive elongation and relieves pauses of transcription. This regulation process depends on the association between 7SK and a HEXIM protein, neither isolated partner being able to inhibit P-TEFb alone. In this work, we used a combined NMR and biochemical approach to determine 7SK and HEXIM1 elements that define their binding properties. Our results demonstrate that a repeated GAUC motif located in the upper part of a hairpin on the 5'-end of 7SK is essential for specific HEXIM1 recognition. Binding of a peptide comprising the HEXIM Arginine Rich Motif (ARM) induces an opening of the GAUC motif and stabilization of an internal loop. A conserved proline-serine sequence in the middle of the ARM is shown to be essential for the binding specificity and the conformational change of the RNA. This work provides evidences for a recognition mechanism involving a first event of induced fit, suggesting that 7SK plasticity is involved in the transcription regulation.
Figures





References
-
- Zieve G, Benecke BJ, Penman S. Synthesis of two classes of small RNA species in vivo and in vitro. Biochemistry. 1977;16:4520–4525. - PubMed
-
- Murphy S, Di Liegro C, Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987;51:81–87. - PubMed
-
- Kruger W, Benecke BJ. Structural and functional analysis of a human 7 S K RNA gene. J. Mol. Biol. 1987;195:31–41. - PubMed

